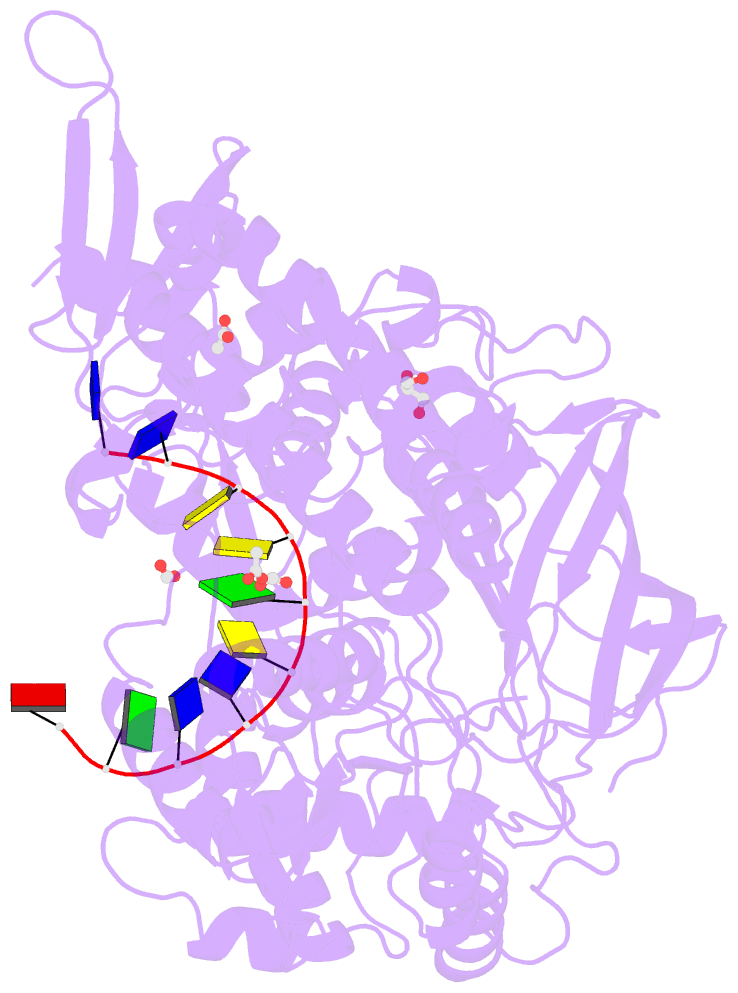
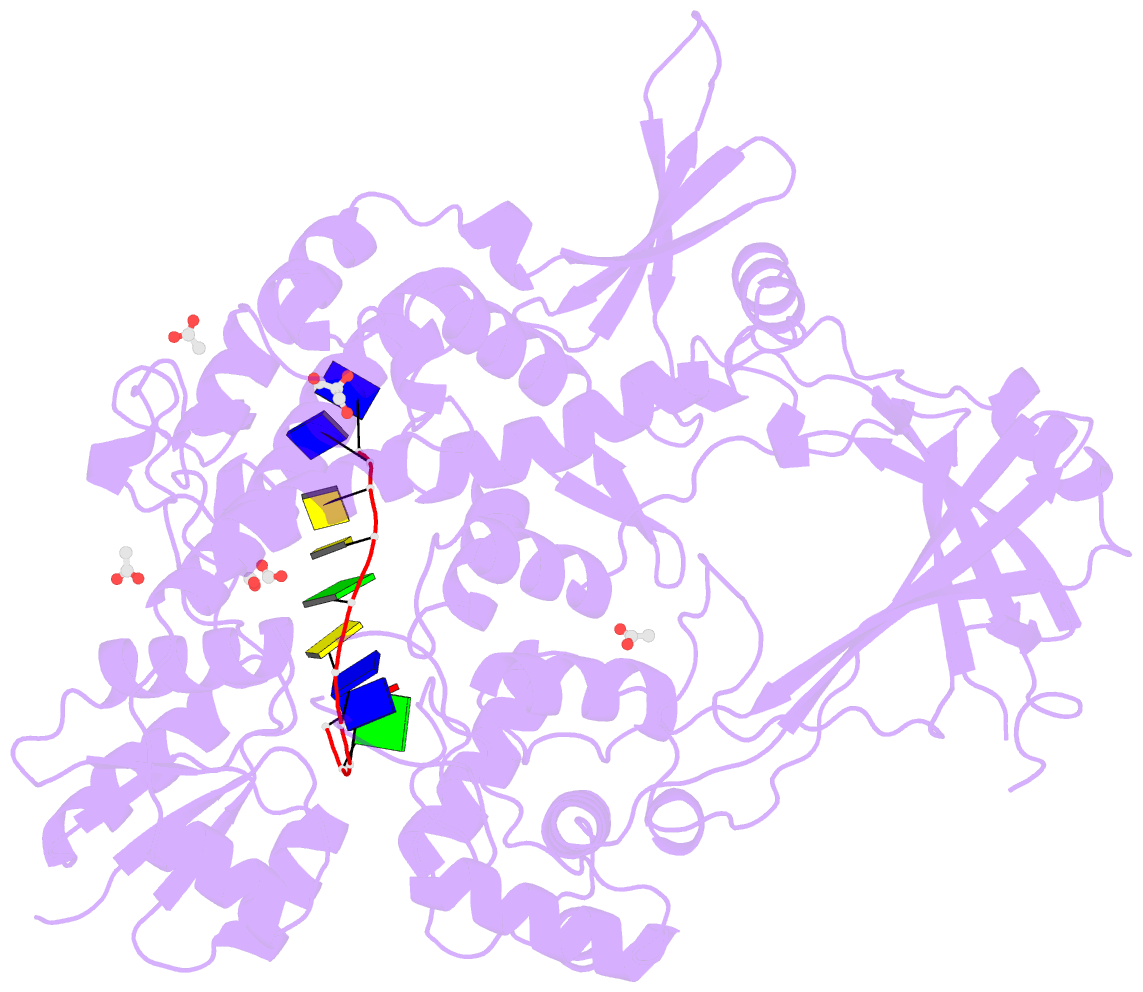
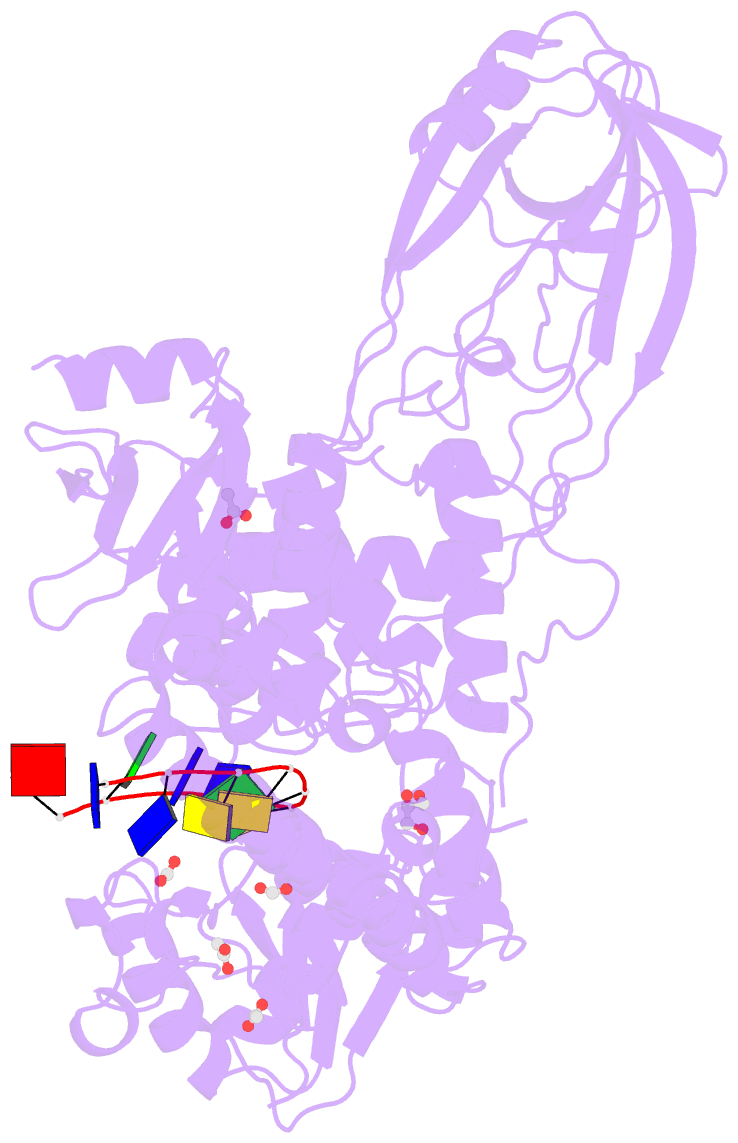
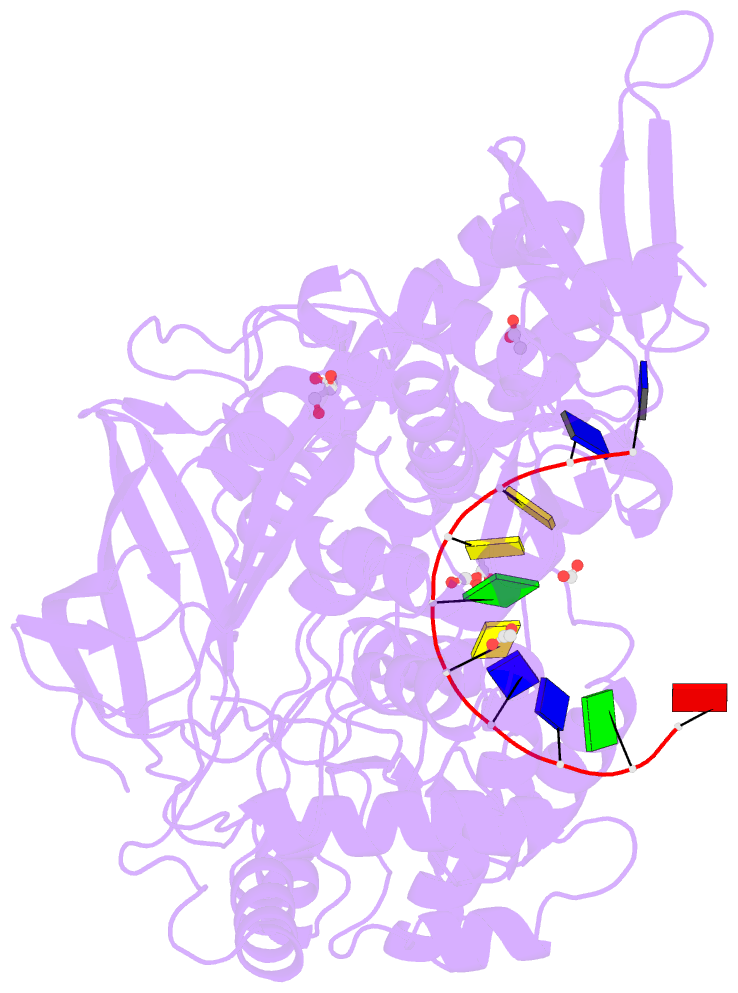
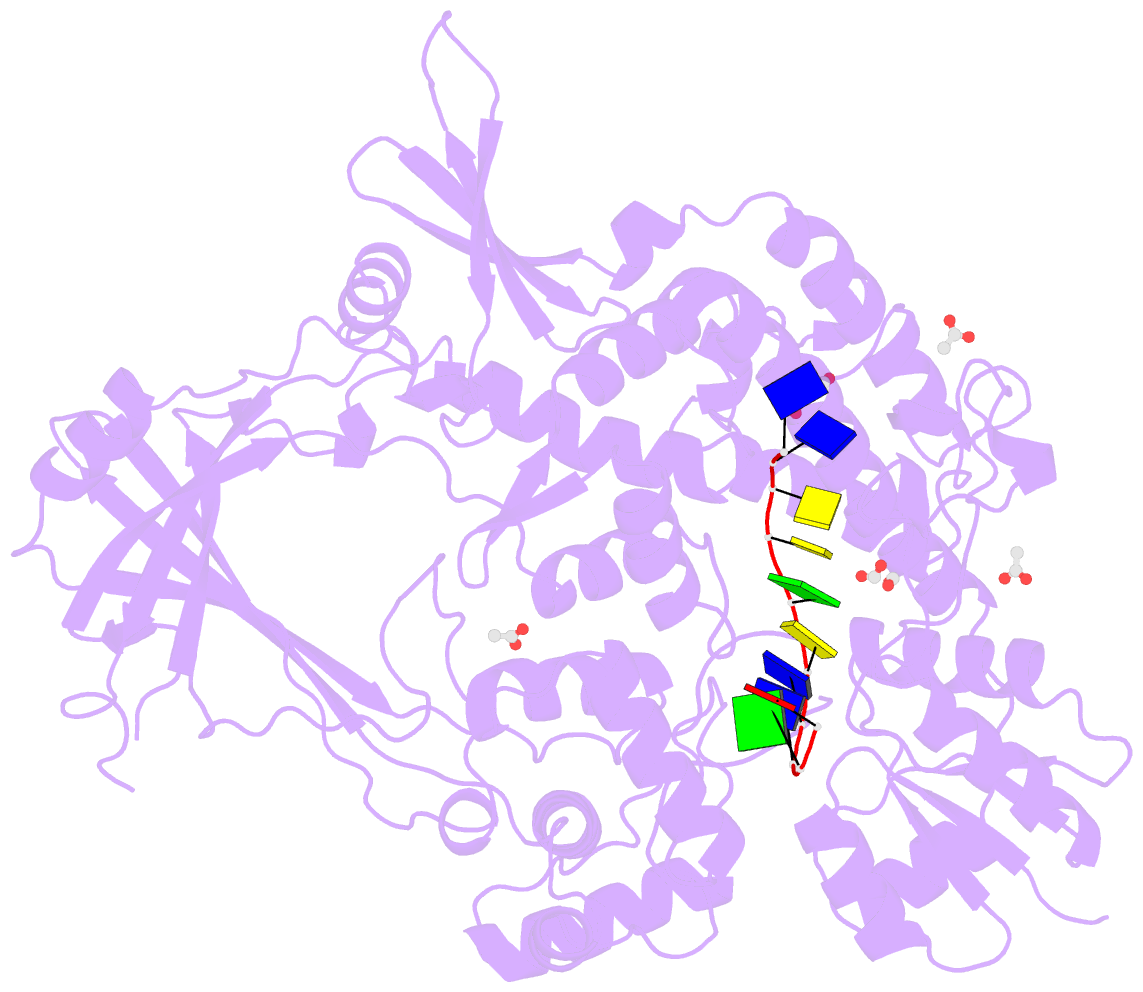
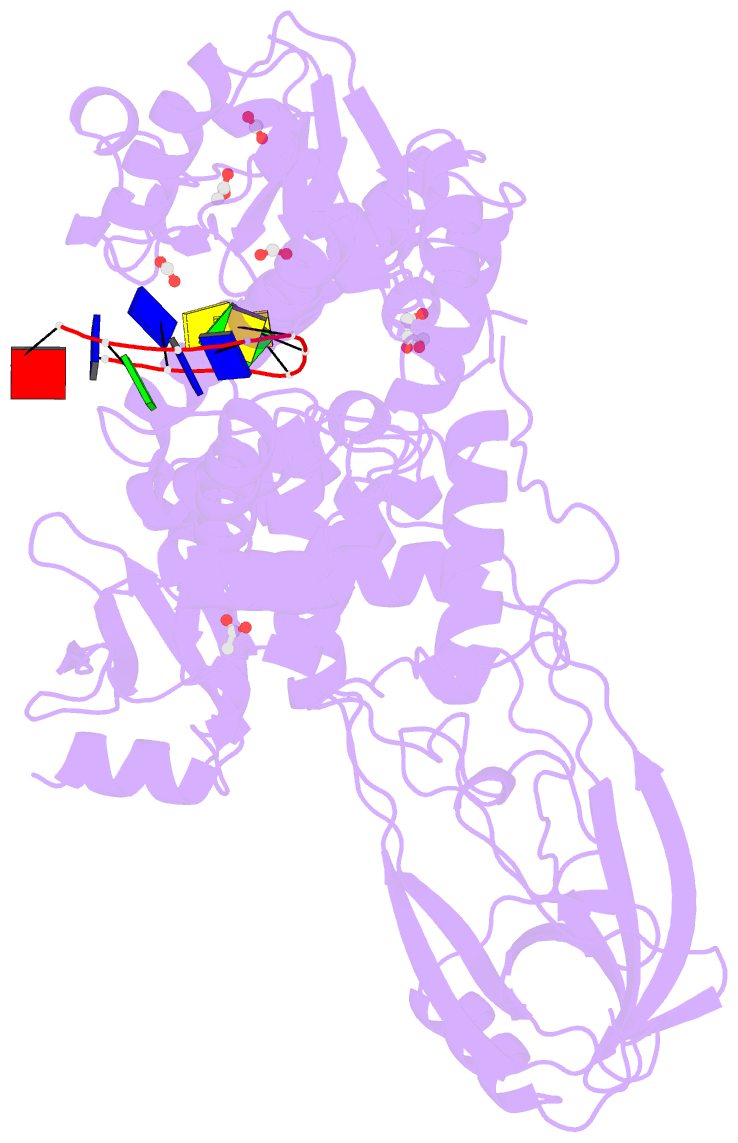
Summary information and primary citation
- PDB-id
- 6cqi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-DNA
- Method
- X-ray (2.42 Å)
- Summary
- 2.42a crystal structure of mycobacterium tuberculosis topoisomerase i in complex with an oligonucleotide mts2-11
- Reference
- Cao N, Tan K, Annamalai T, Joachimiak A, Tse-Dinh YC (2018): "Investigating mycobacterial topoisomerase I mechanism from the analysis of metal and DNA substrate interactions at the active site." Nucleic Acids Res., 46, 7296-7308. doi: 10.1093/nar/gky492.
- Abstract
- We have obtained new crystal structures of Mycobacterium tuberculosis topoisomerase I, including structures with ssDNA substrate bound to the active site, with and without Mg2+ ion present. Significant enzyme conformational changes upon DNA binding place the catalytic tyrosine in a pre-transition state position for cleavage of a specific phosphodiester linkage. Meanwhile, the enzyme/DNA complex with bound Mg2+ ion may represent the post-transition state for religation in the enzyme's multiple-step DNA relaxation catalytic cycle. The first observation of Mg2+ ion coordinated with the TOPRIM residues and DNA phosphate in a type IA topoisomerase active site allows assignment of likely catalytic role for the metal and draws a comparison to the proposed mechanism for type IIA topoisomerases. The critical function of a strictly conserved glutamic acid in the DNA cleavage step was assessed through site-directed mutagenesis. The functions assigned to the observed Mg2+ ion can account for the metal requirement for DNA rejoining but not DNA cleavage by type IA topoisomerases. This work provides new structural insights into a more stringent requirement for DNA rejoining versus cleavage in the catalytic cycle of this essential enzyme, and further establishes the potential for selective interference of DNA rejoining by this validated TB drug target.