Summary information and primary citation
- PDB-id
- 6cst; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication, transferase-DNA
- Method
- X-ray (2.0 Å)
- Summary
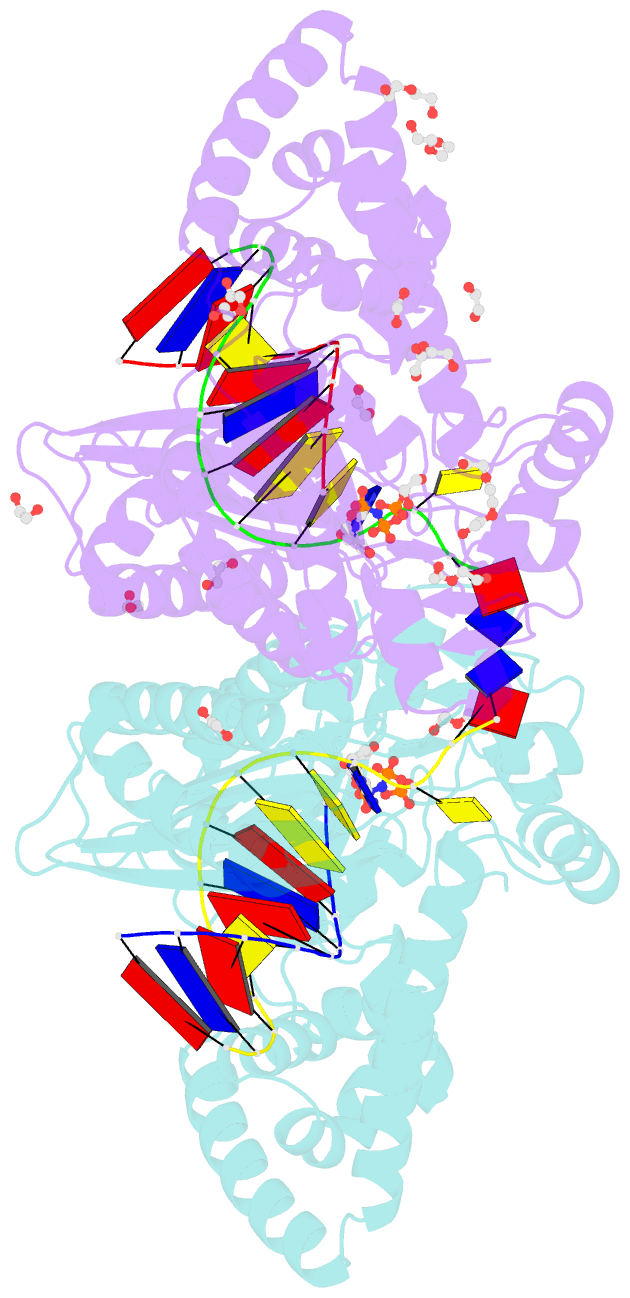
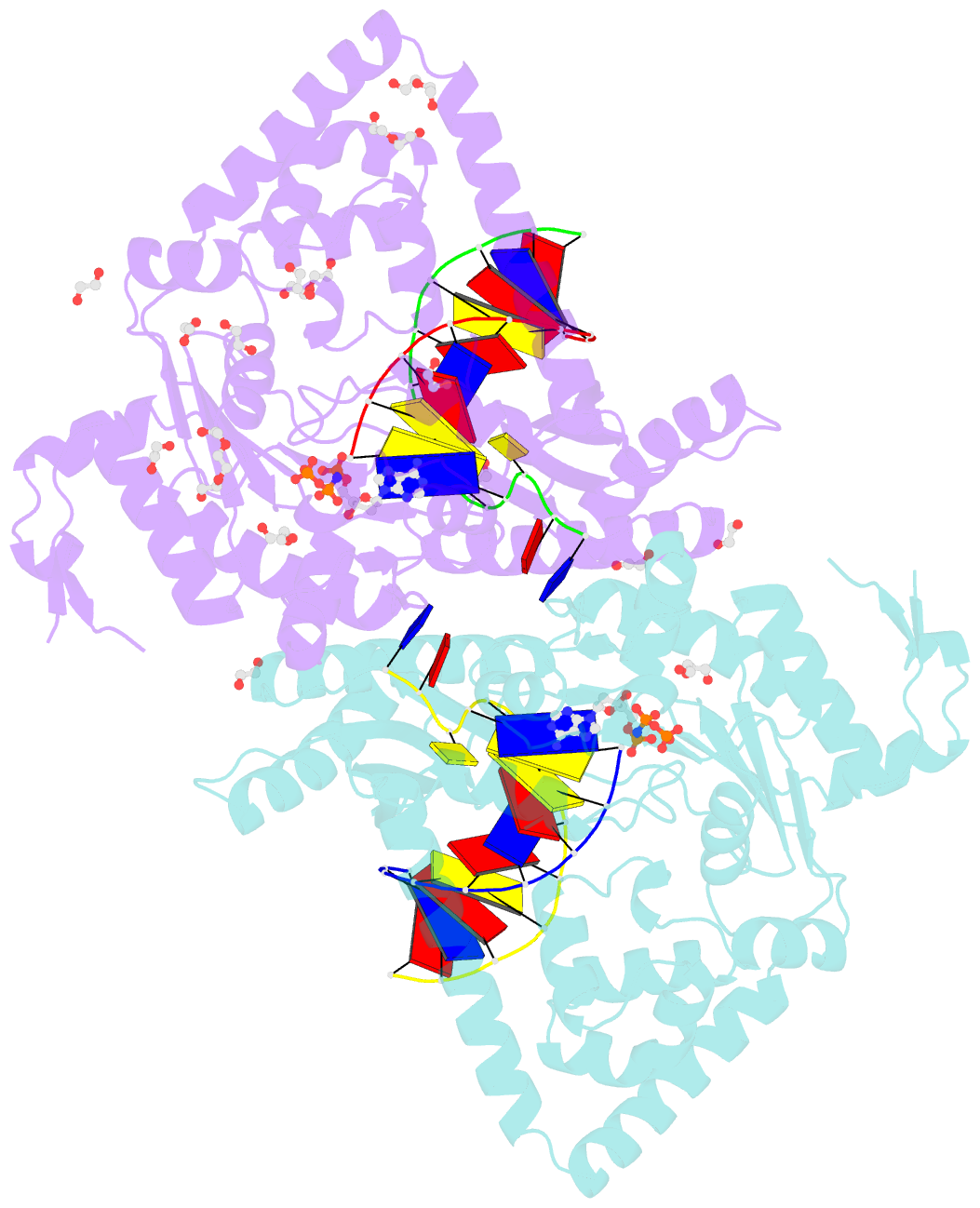
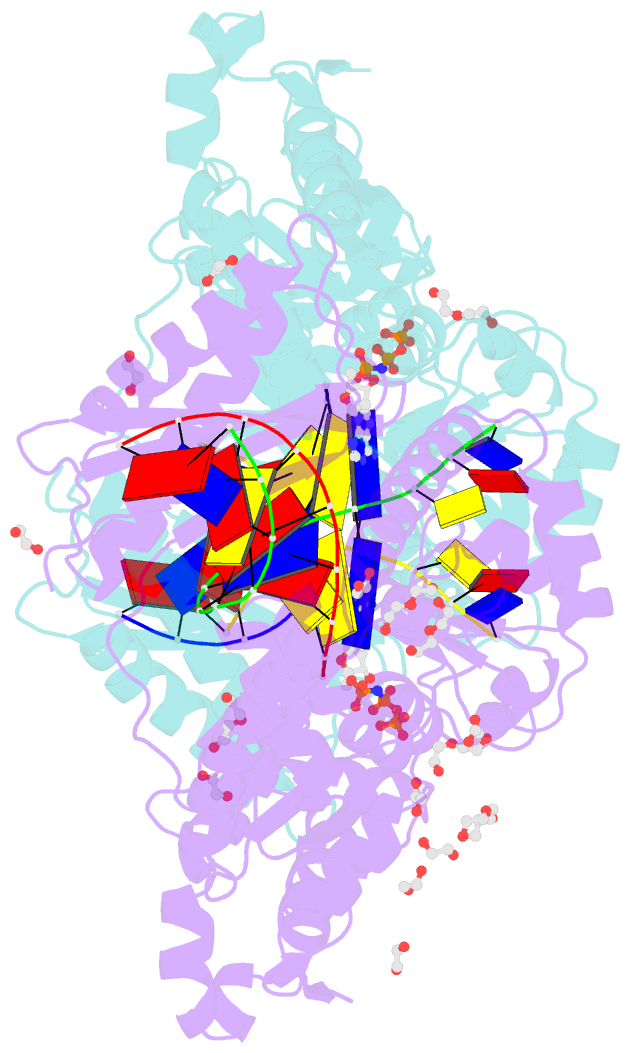
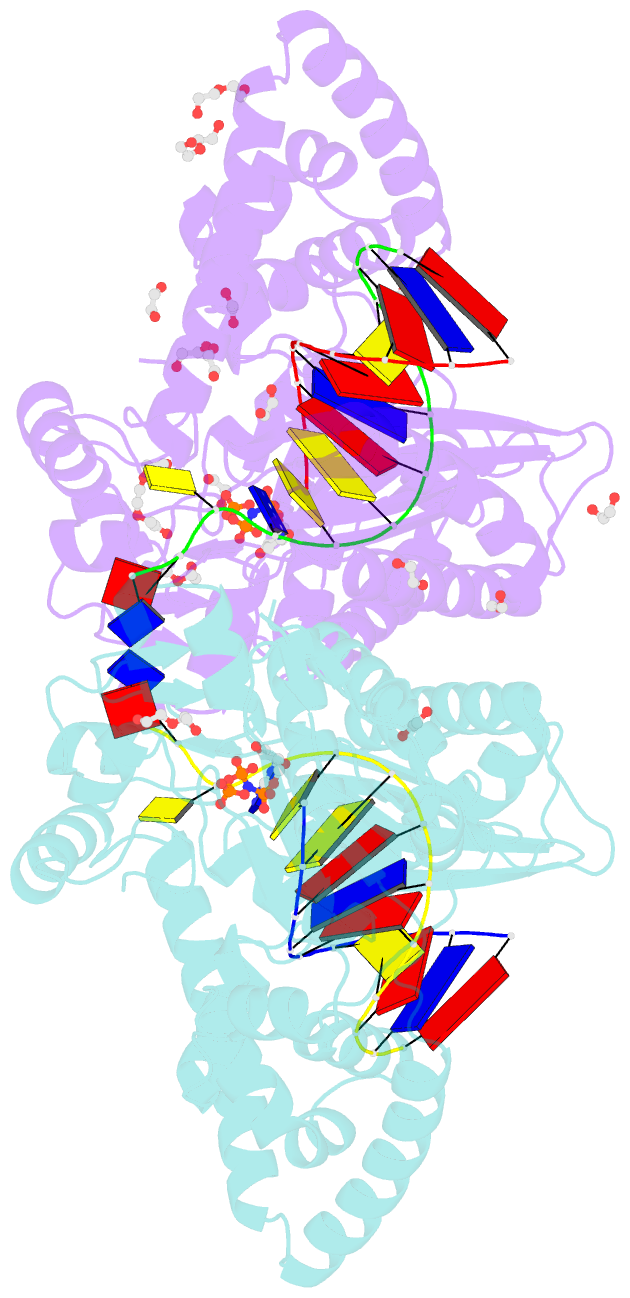
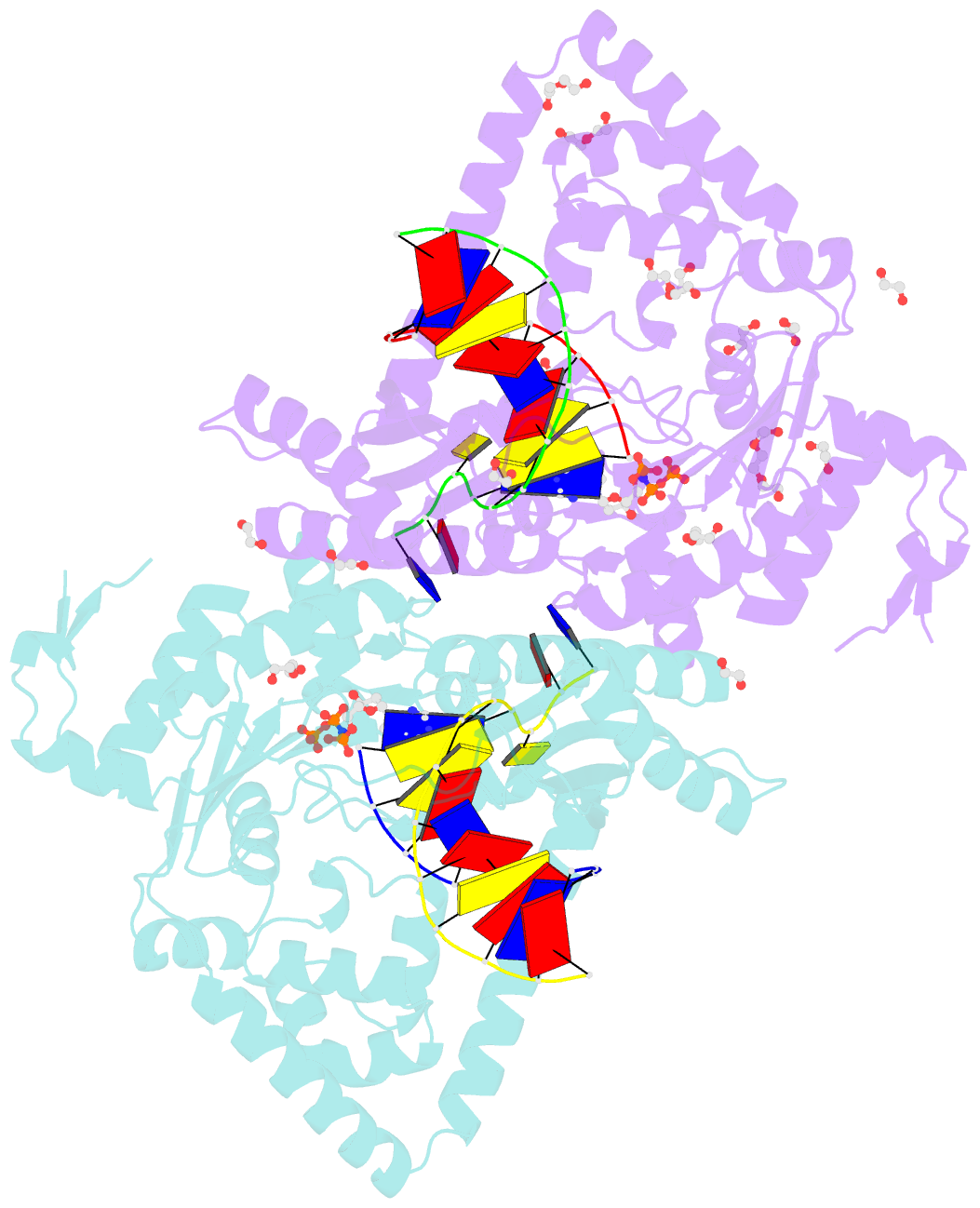
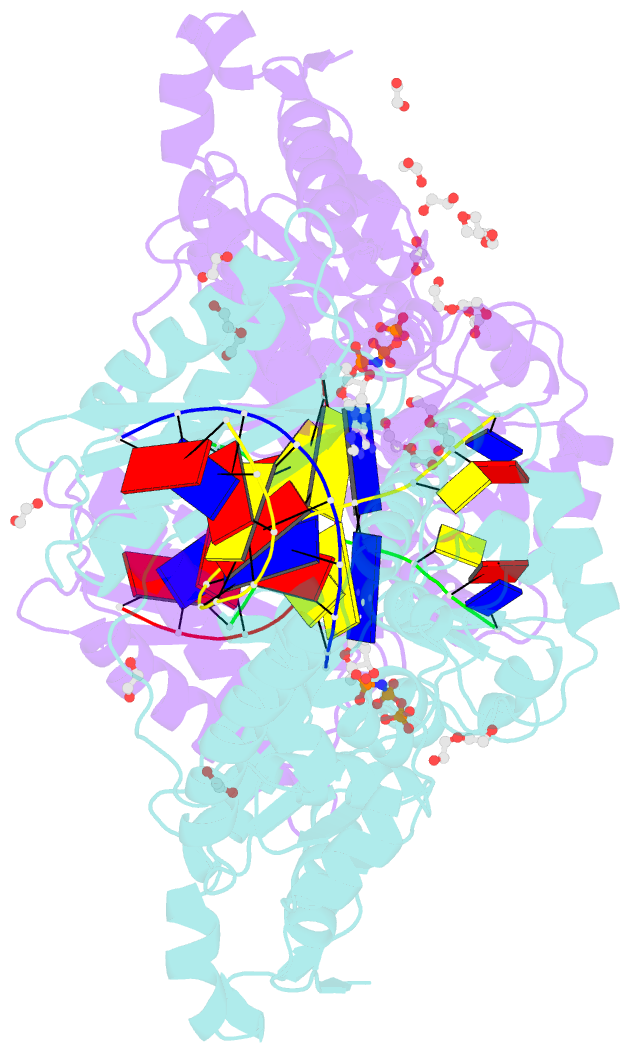
- Structure of human DNA polymerase kappa with DNA
- Reference
- Jha V, Ling H (2018): "2.0 angstrom resolution crystal structure of human pol kappa reveals a new catalytic function of N-clasp in DNA replication." Sci Rep, 8, 15125. doi: 10.1038/s41598-018-33371-5.
- Abstract
- Human polymerase kappa (polκ) is a distinct Y-family DNA polymerase with a unique N-terminal N-clasp domain. The N-clasp renders polκ's high efficiency and accuracy in DNA replication and lesion bypass. How N-clasp empowers polκ in replication remains unclear due to the disordering of N-clasp. Here, we present a 2.0-Å resolution crystal structure of a polκ ternary complex with DNA and an incoming nucleotide. The structure-function study reveals an ordered N-clasp domain that brings conserved and functionally important residues in contact with the replicating basepair in the active site and contributes to the nucleotidyl transfer reaction. Particularly, a fully ordered Lys25 from the N-clasp domain is in H-bonding with the α- and γ-phosphates of the incoming nucleotide. K25A mutation reduces the polymerase activity of polκ significantly. This lysine is structurally analogous to a conserved lysine in the A-family DNA polymerases in the closed form. In contrast, Lys25 in the previous structures of polκ does not have any contacts with the incoming nucleotide, resembling an open form of a DNA polymerase. Based on structural and functional similarity, we propose a local open/closed mechanism for polκ in DNA replication catalysis, which mimics the common mechanism for all DNA polymerases.