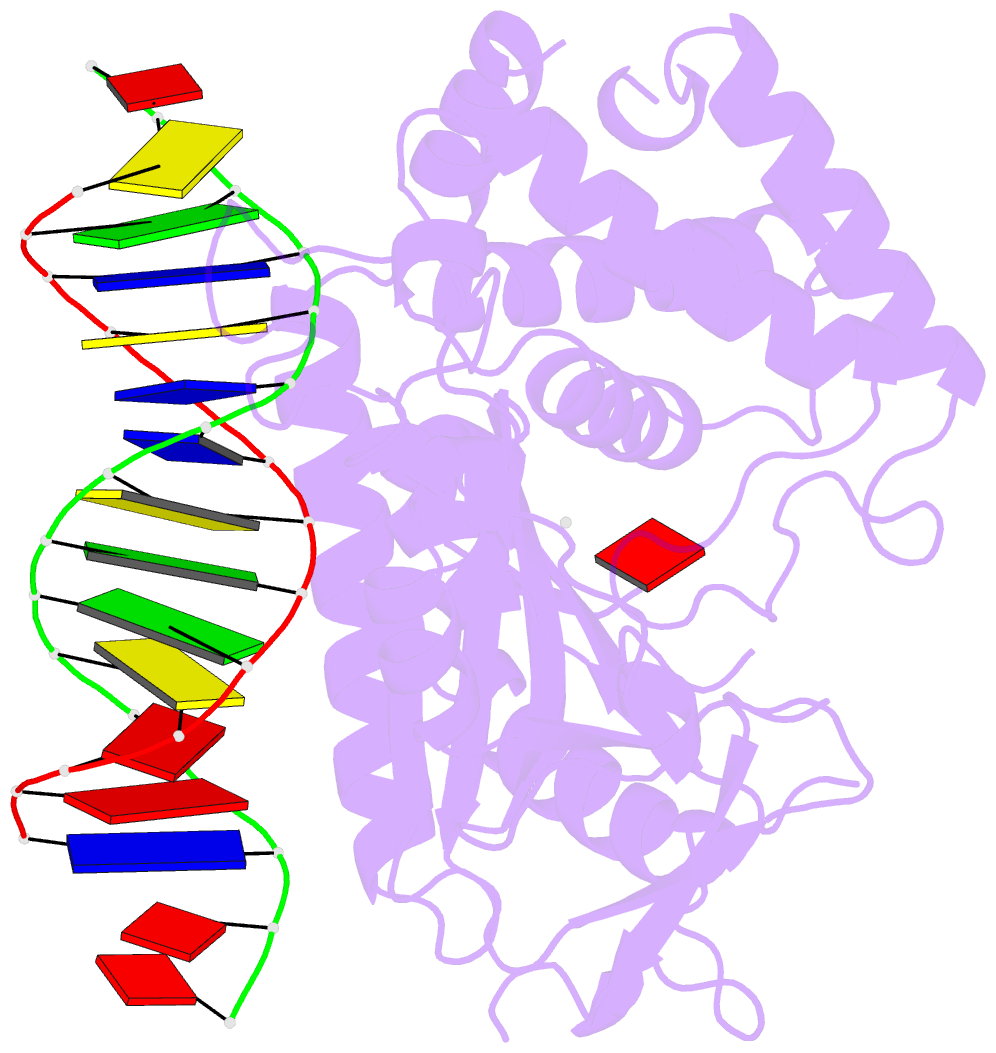
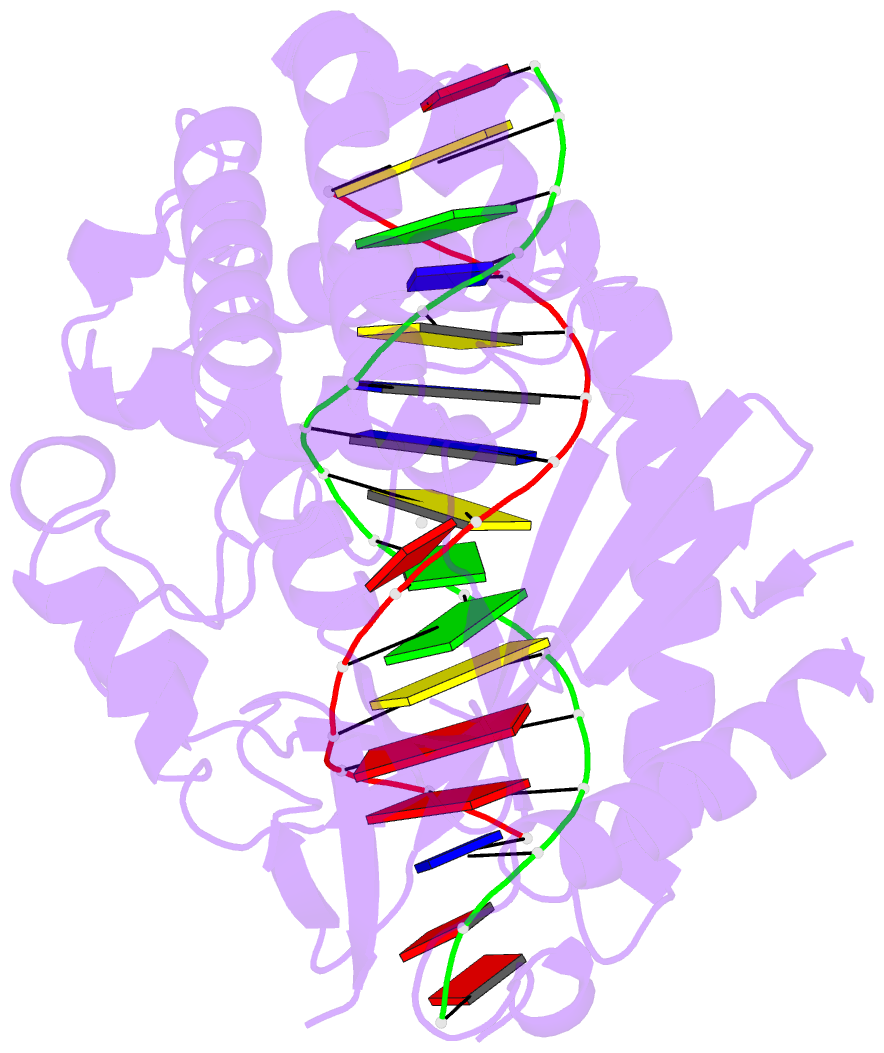
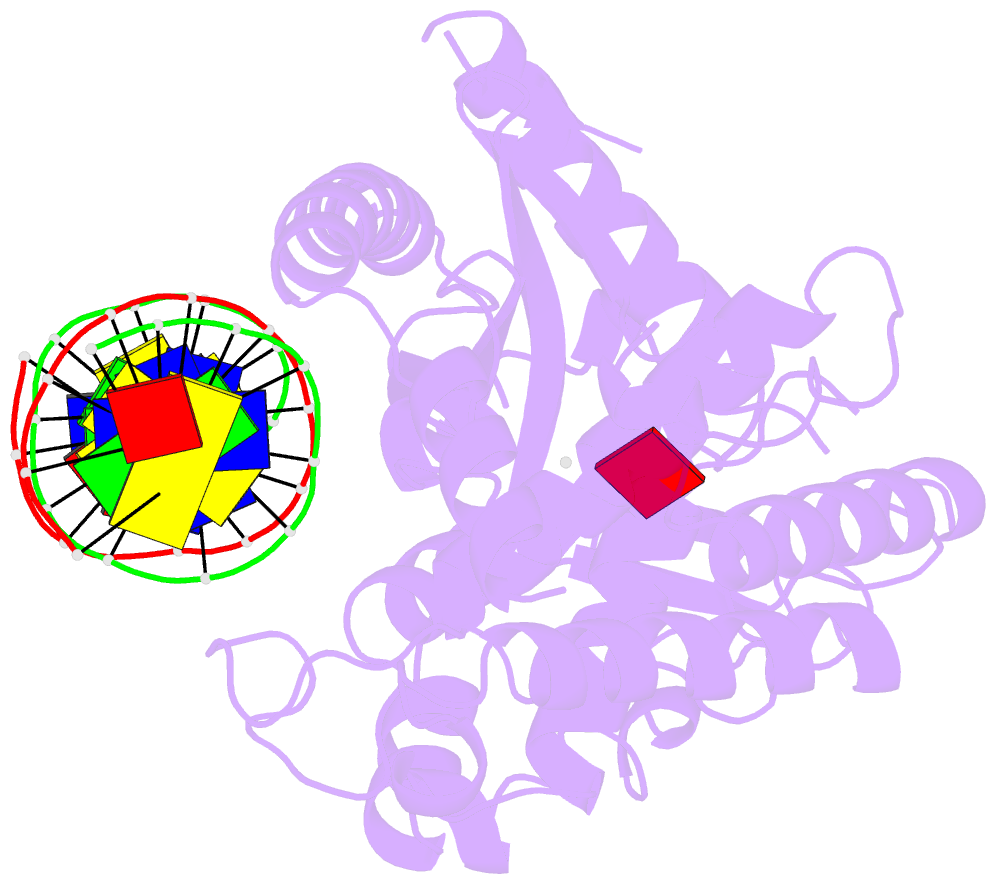
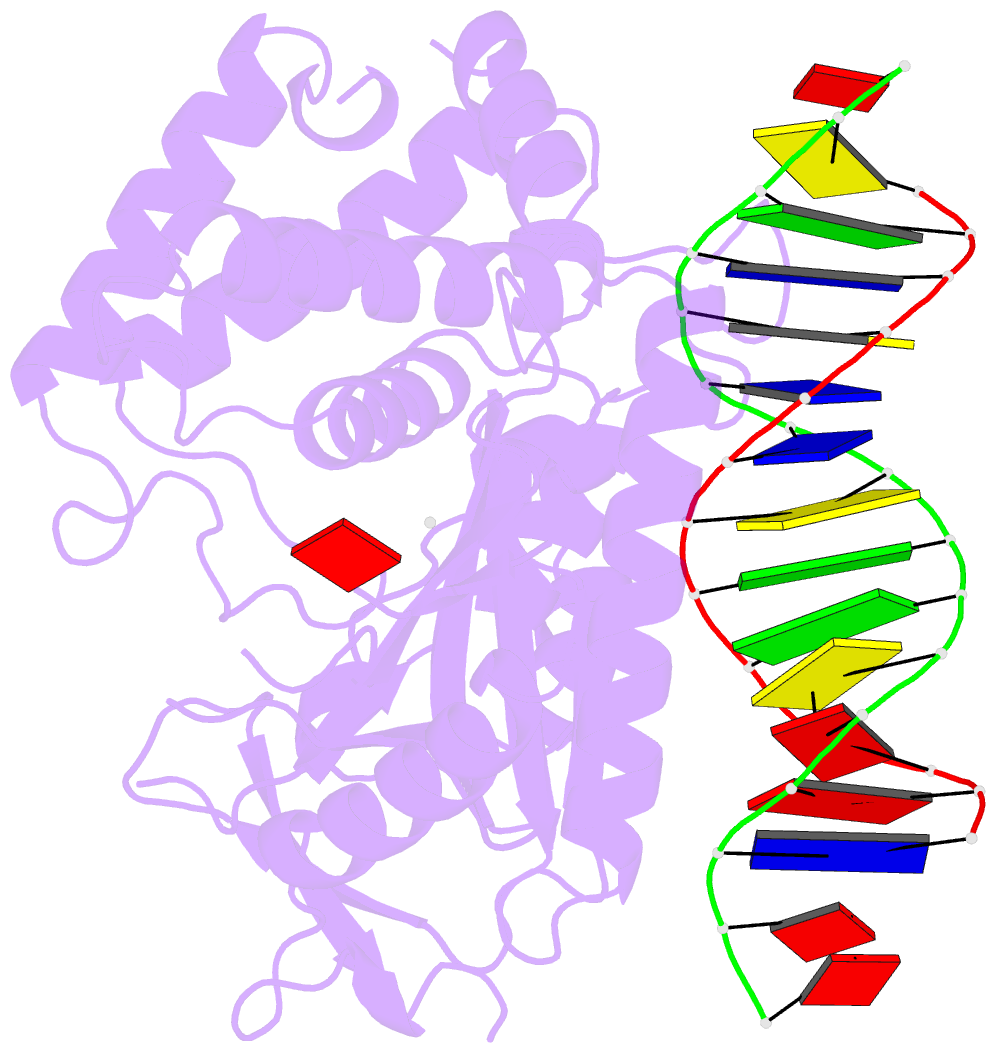
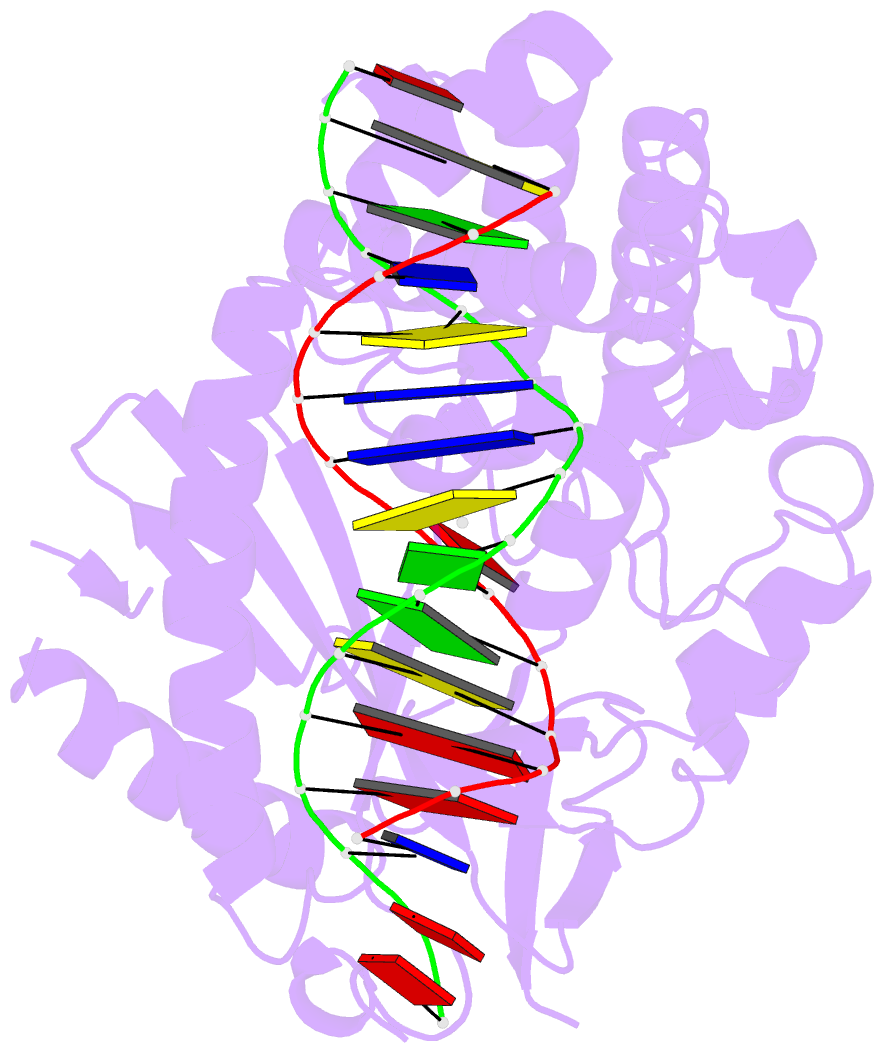
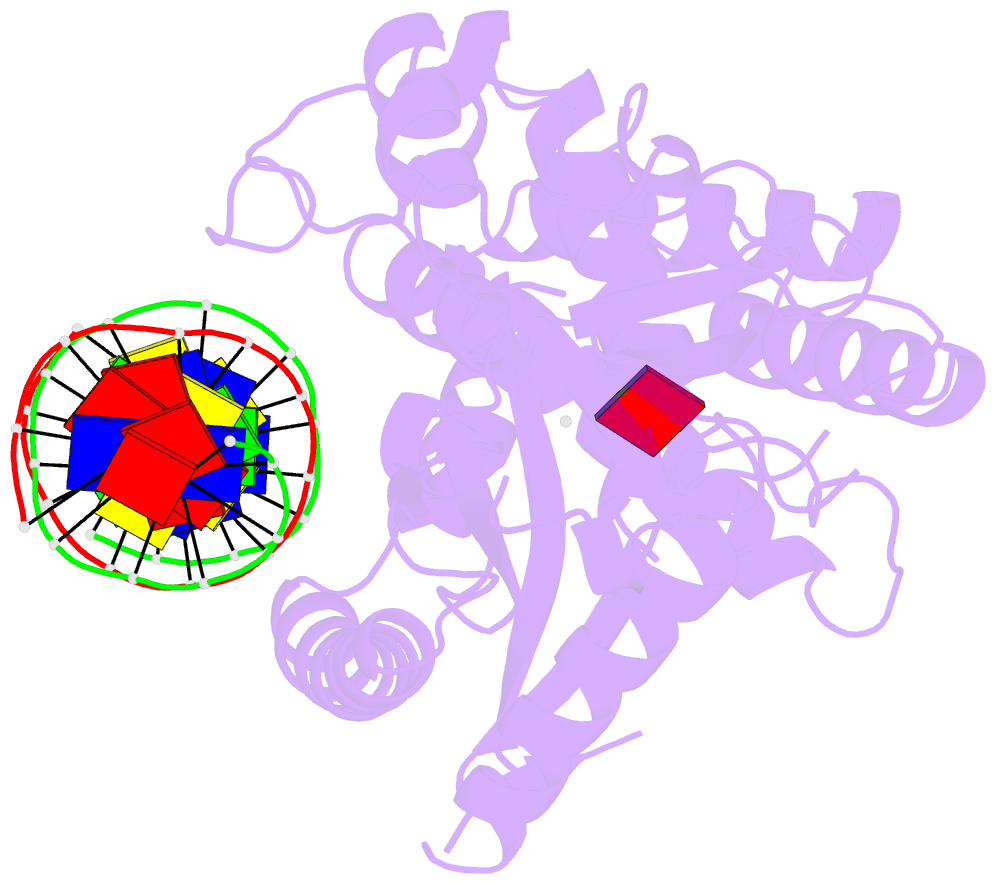
Summary information and primary citation
- PDB-id
- 6cta; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.779 Å)
- Summary
- Structure of the human cgas-DNA complex with atp
- Reference
- Zhou W, Whiteley AT, de Oliveira Mann CC, Morehouse BR, Nowak RP, Fischer ES, Gray NS, Mekalanos JJ, Kranzusch PJ (2018): "Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance." Cell, 174, 300-311.e11. doi: 10.1016/j.cell.2018.06.026.
- Abstract
- Cyclic GMP-AMP synthase (cGAS) recognition of cytosolic DNA is critical for immune responses to pathogen replication, cellular stress, and cancer. Existing structures of the mouse cGAS-DNA complex provide a model for enzyme activation but do not explain why human cGAS exhibits severely reduced levels of cyclic GMP-AMP (cGAMP) synthesis compared to other mammals. Here, we discover that enhanced DNA-length specificity restrains human cGAS activation. Using reconstitution of cGAMP signaling in bacteria, we mapped the determinant of human cGAS regulation to two amino acid substitutions in the DNA-binding surface. Human-specific substitutions are necessary and sufficient to direct preferential detection of long DNA. Crystal structures reveal why removal of human substitutions relaxes DNA-length specificity and explain how human-specific DNA interactions favor cGAS oligomerization. These results define how DNA-sensing in humans adapted for enhanced specificity and provide a model of the active human cGAS-DNA complex to enable structure-guided design of cGAS therapeutics.