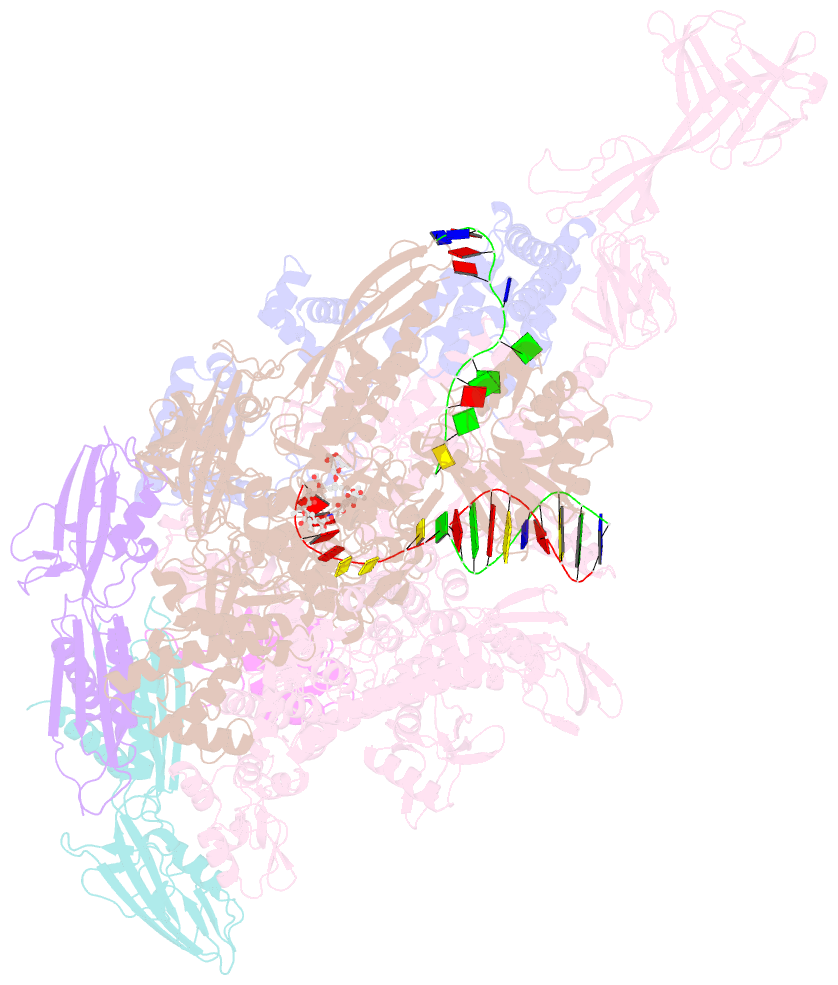
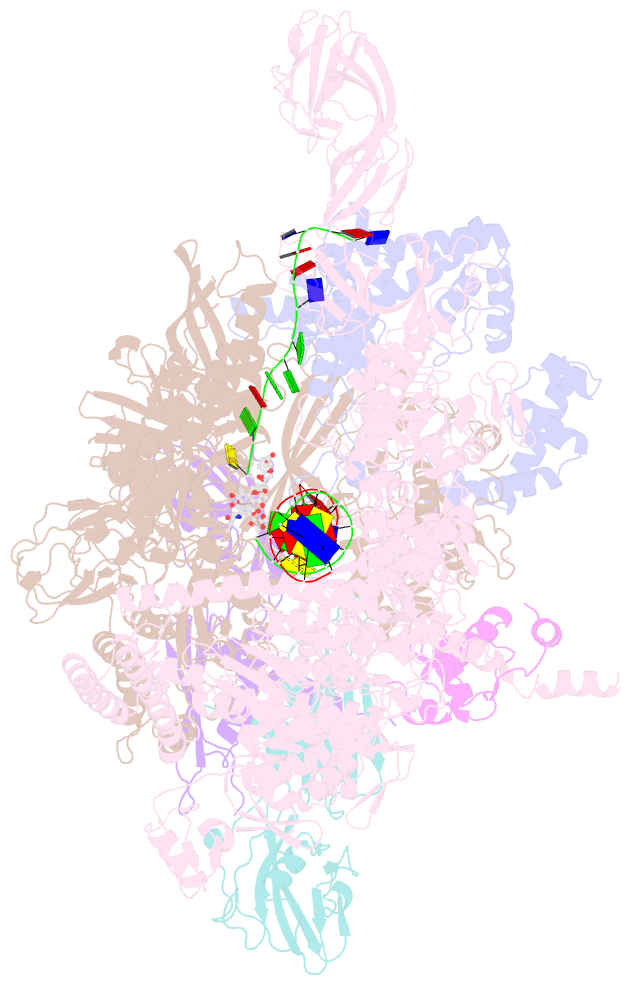
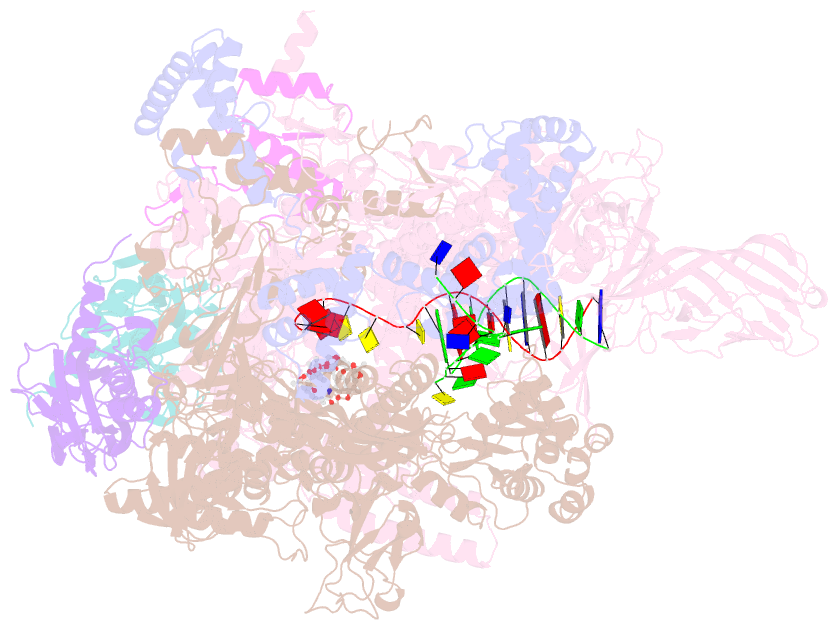
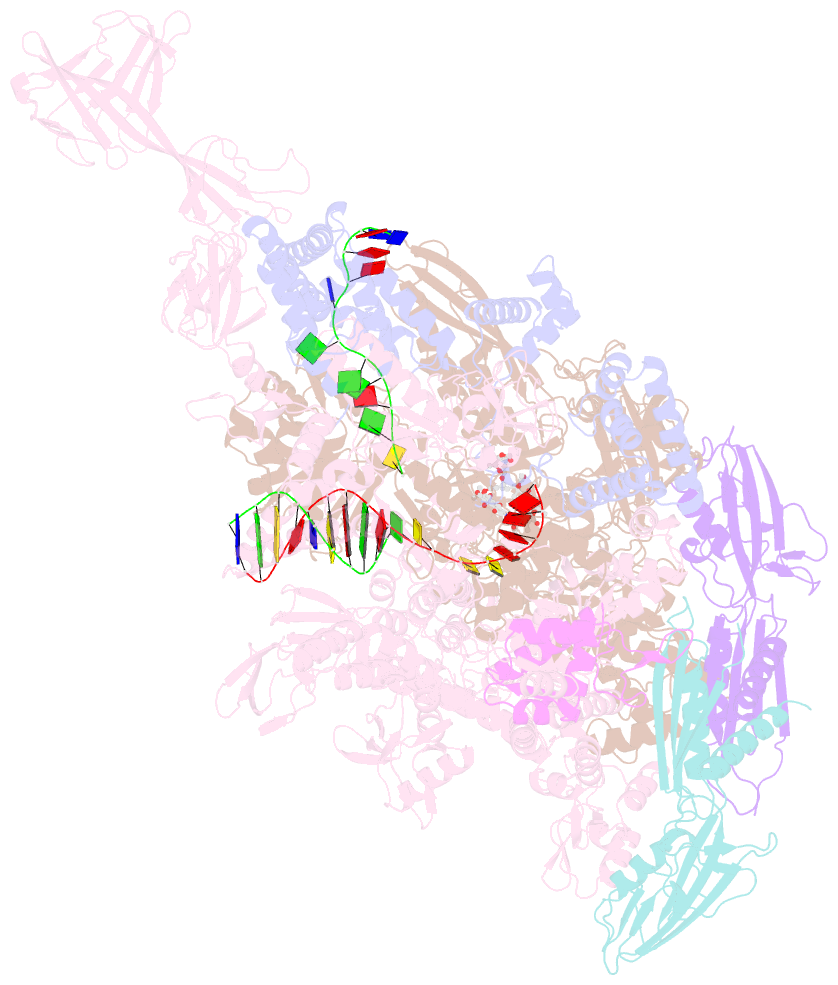
Summary information and primary citation
- PDB-id
- 6cuu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA-antibiotic
- Method
- X-ray (2.994 Å)
- Summary
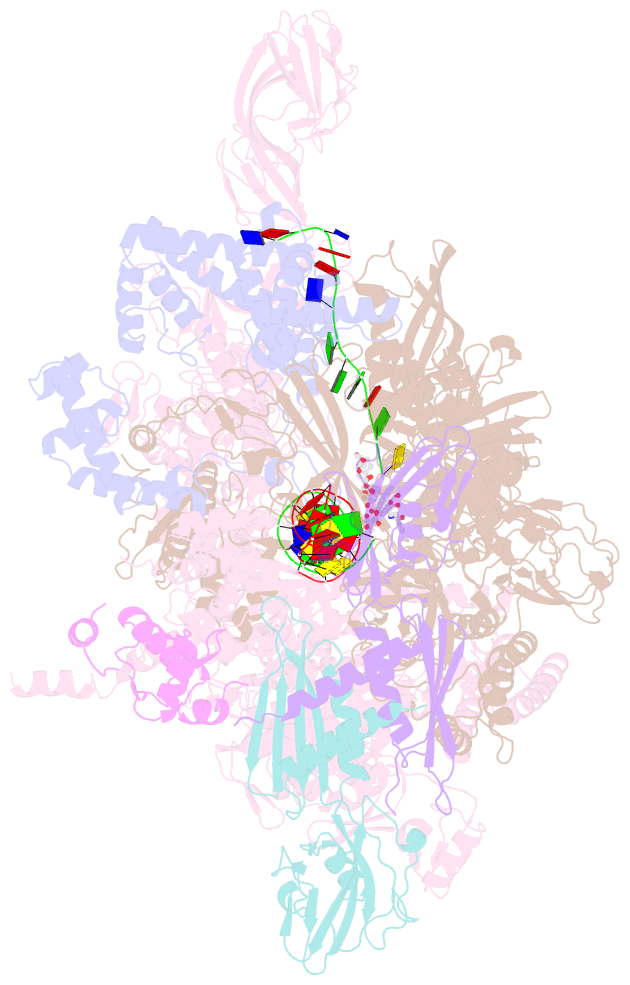
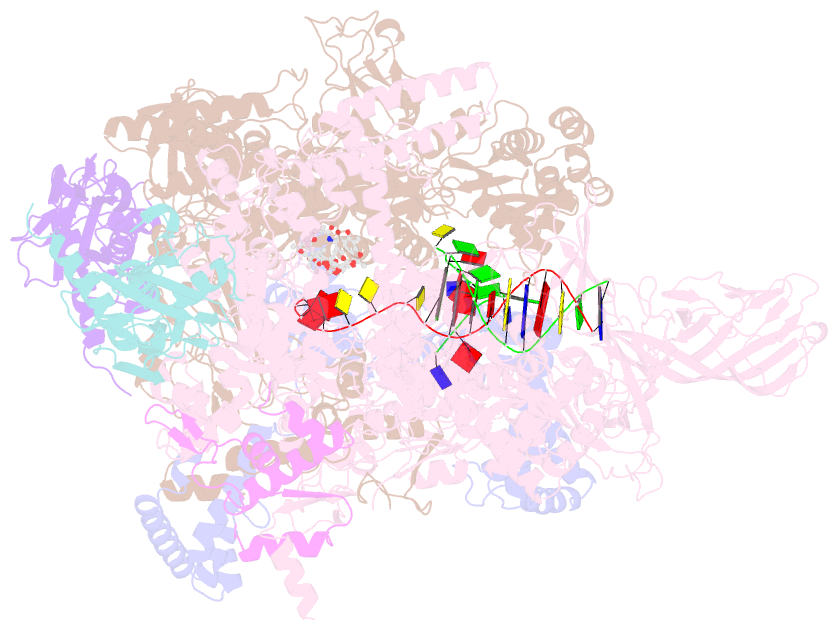
- Thermus thermophiles RNA polymerase in complex with promoter DNA and antibiotic kanglemycin a
- Reference
- Mosaei H, Molodtsov V, Kepplinger B, Harbottle J, Moon CW, Jeeves RE, Ceccaroni L, Shin Y, Morton-Laing S, Marrs ECL, Wills C, Clegg W, Yuzenkova Y, Perry JD, Bacon J, Errington J, Allenby NEE, Hall MJ, Murakami KS, Zenkin N (2018): "Mode of Action of Kanglemycin A, an Ansamycin Natural Product that Is Active against Rifampicin-Resistant Mycobacterium tuberculosis." Mol. Cell, 72, 263-274.e5. doi: 10.1016/j.molcel.2018.08.028.
- Abstract
- Antibiotic-resistant bacterial pathogens pose an urgent healthcare threat, prompting a demand for new medicines. We report the mode of action of the natural ansamycin antibiotic kanglemycin A (KglA). KglA binds bacterial RNA polymerase at the rifampicin-binding pocket but maintains potency against RNA polymerases containing rifampicin-resistant mutations. KglA has antibiotic activity against rifampicin-resistant Gram-positive bacteria and multidrug-resistant Mycobacterium tuberculosis (MDR-M. tuberculosis). The X-ray crystal structures of KglA with the Escherichia coli RNA polymerase holoenzyme and Thermus thermophilus RNA polymerase-promoter complex reveal an altered-compared with rifampicin-conformation of KglA within the rifampicin-binding pocket. Unique deoxysugar and succinate ansa bridge substituents make additional contacts with a separate, hydrophobic pocket of RNA polymerase and preclude the formation of initial dinucleotides, respectively. Previous ansa-chain modifications in the rifamycin series have proven unsuccessful. Thus, KglA represents a key starting point for the development of a new class of ansa-chain derivatized ansamycins to tackle rifampicin resistance.