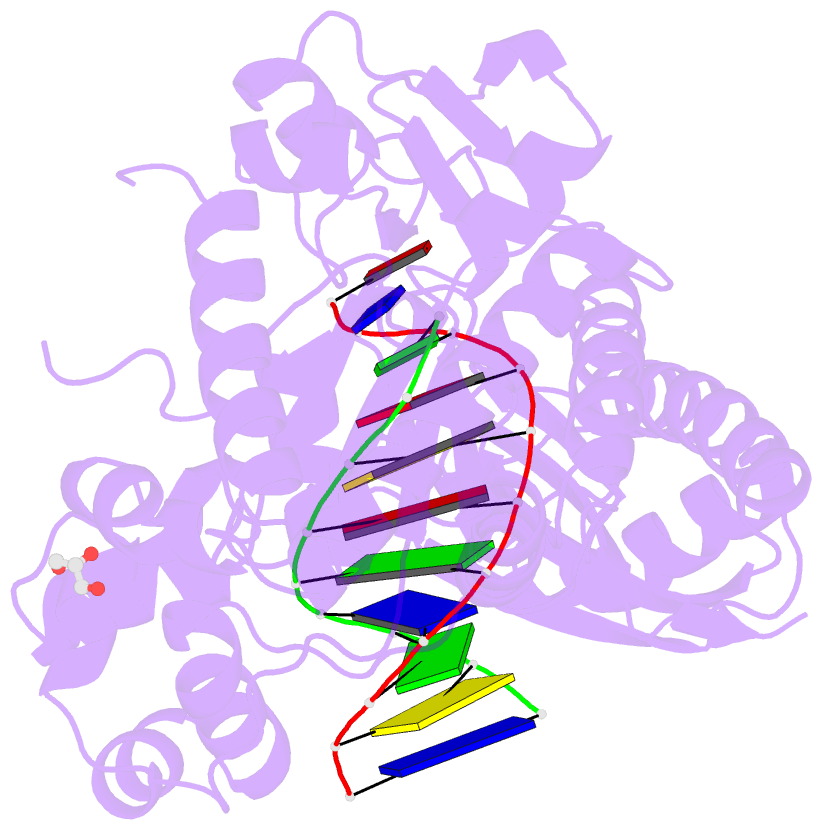
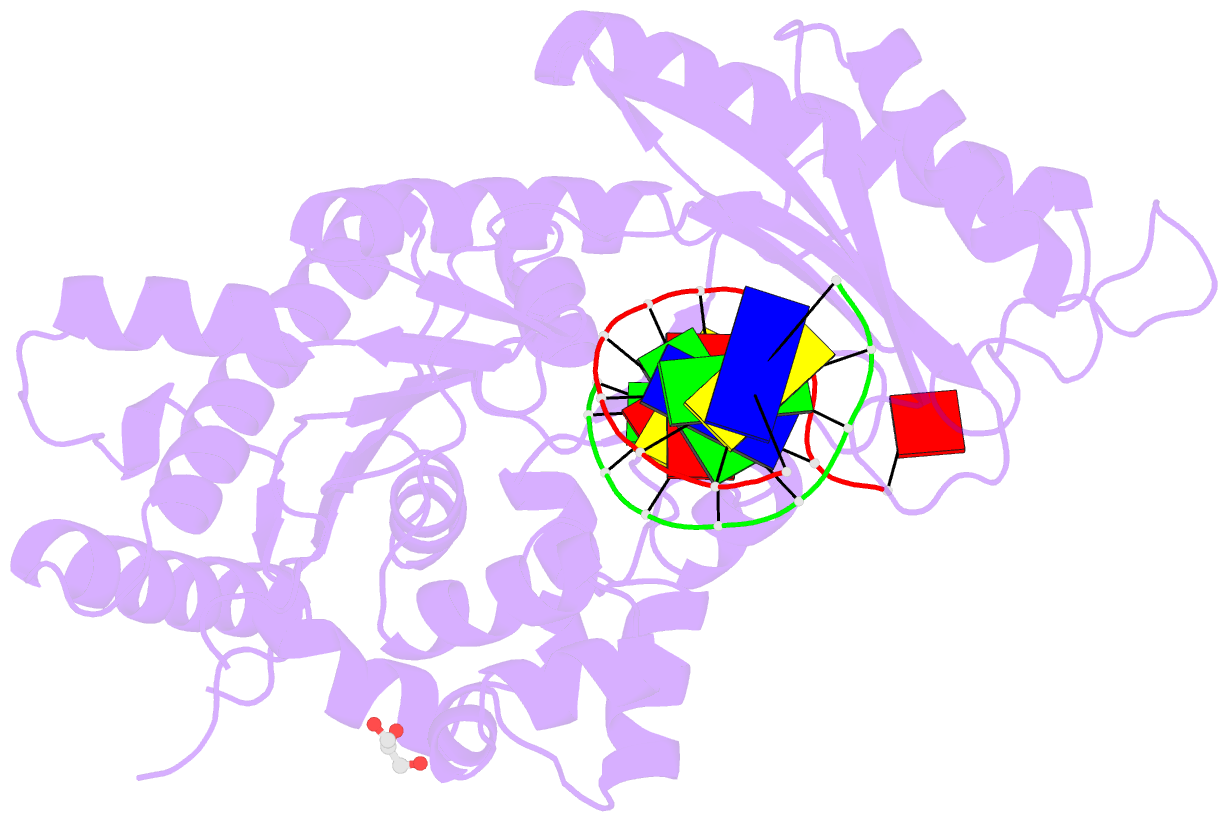
Summary information and primary citation
- PDB-id
- 6d0m; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.832 Å)
- Summary
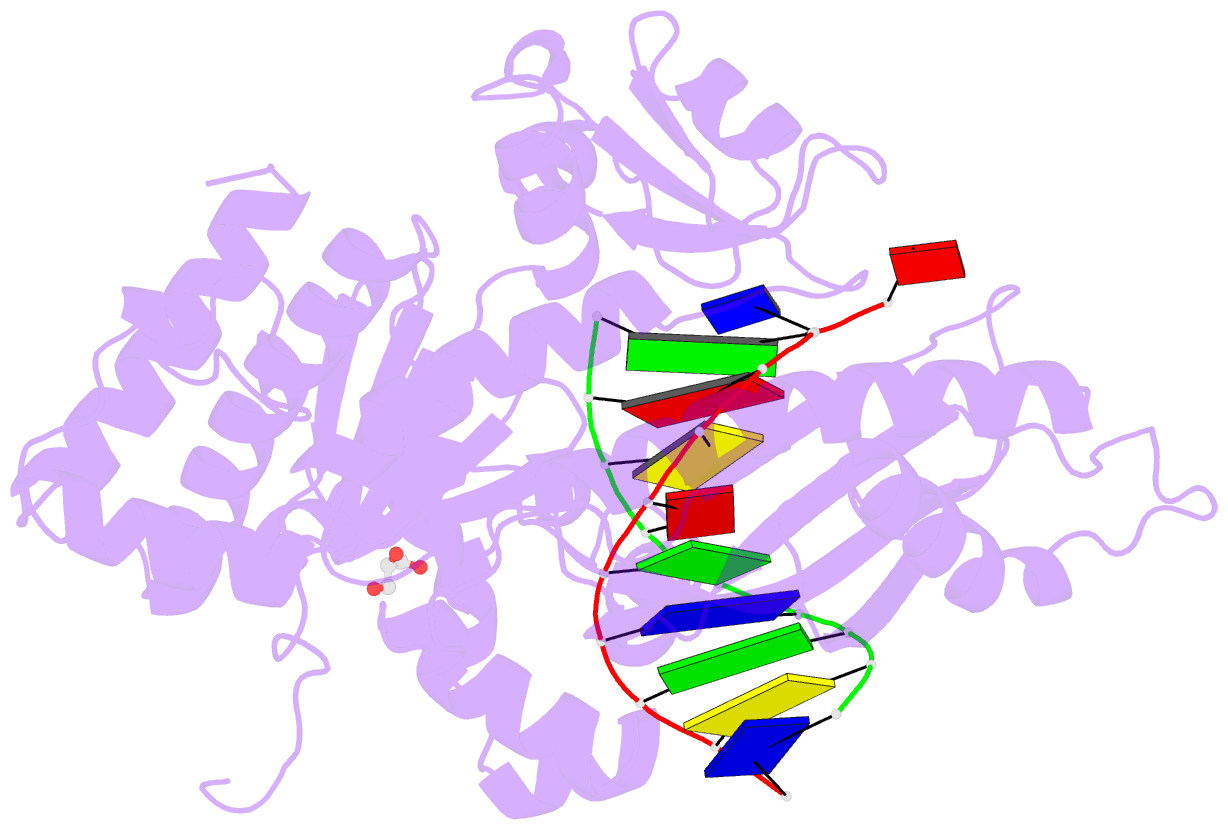
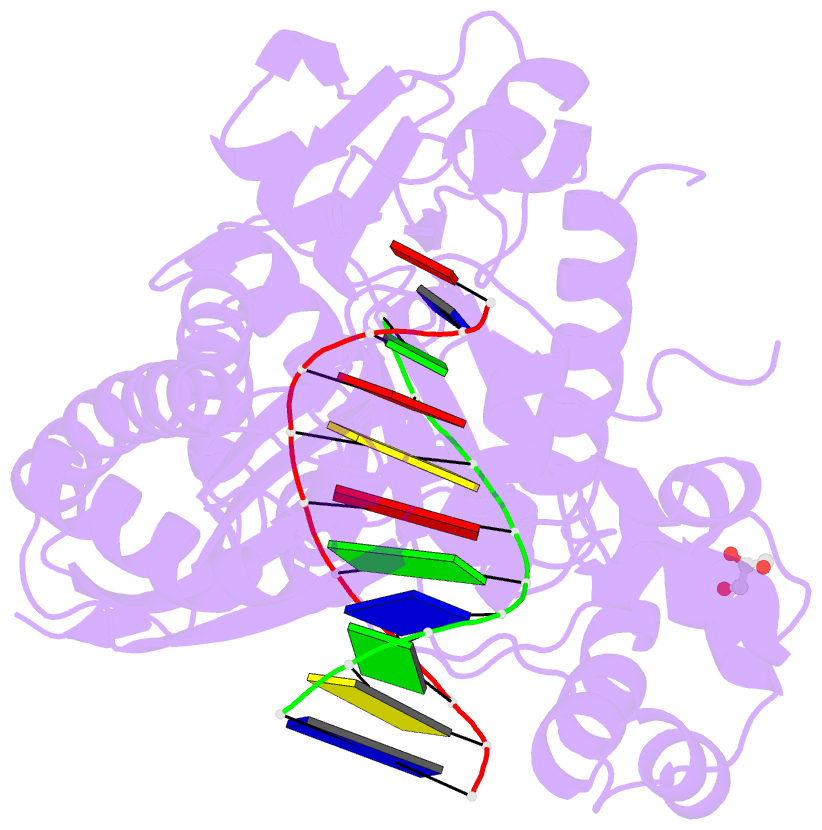
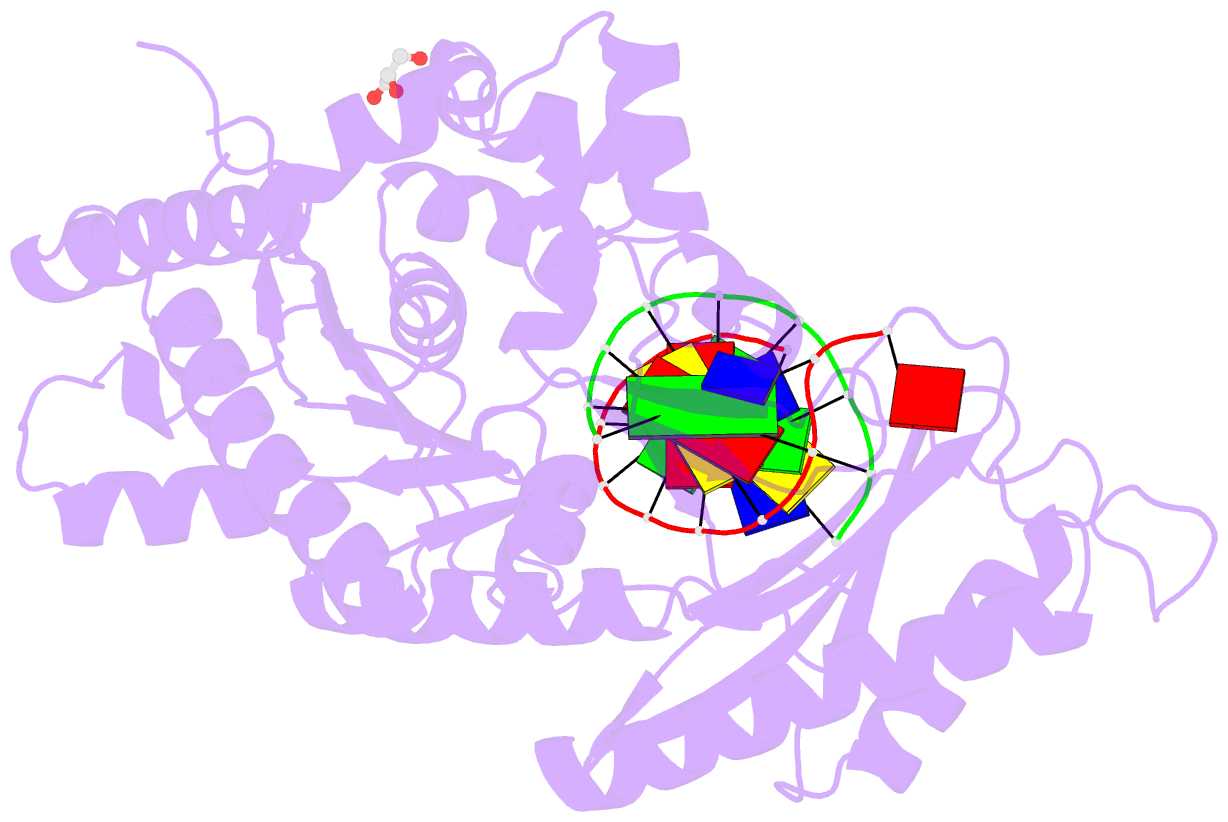
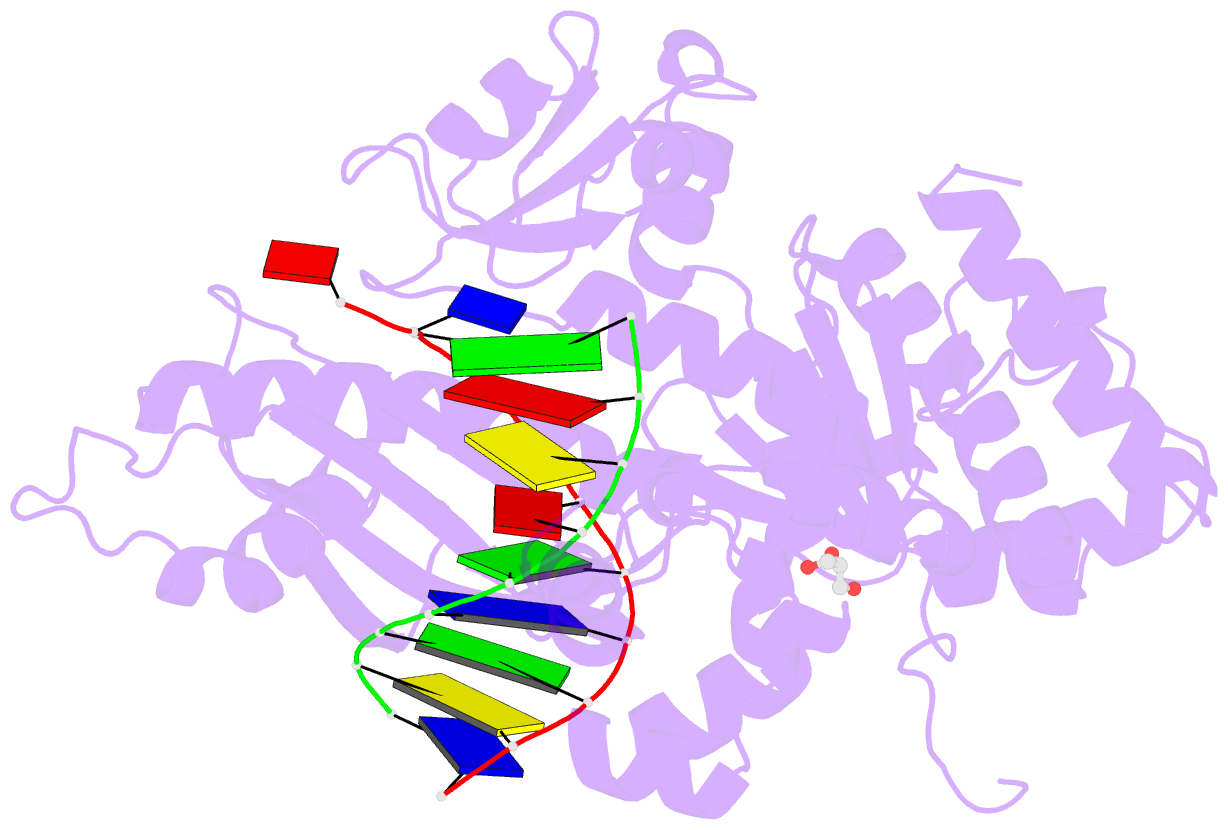
- Polymerase eta post-insertion binary complex with cytarabine (arac)
- Reference
- Rechkoblit O, Choudhury JR, Buku A, Prakash L, Prakash S, Aggarwal AK (2018): "Structural basis for polymerase eta-promoted resistance to the anticancer nucleoside analog cytarabine." Sci Rep, 8, 12702. doi: 10.1038/s41598-018-30796-w.
- Abstract
- Cytarabine (AraC) is an essential chemotherapeutic for acute myeloid leukemia (AML) and resistance to this drug is a major cause of treatment failure. AraC is a nucleoside analog that differs from 2'-deoxycytidine only by the presence of an additional hydroxyl group at the C2' position of the 2'-deoxyribose. The active form of the drug AraC 5'-triphosphate (AraCTP) is utilized by human replicative DNA polymerases to insert AraC at the 3' terminus of a growing DNA chain. This impedes further primer extension and is a primary basis for the drug action. The Y-family translesion synthesis (TLS) DNA polymerase η (Polη) counteracts this barrier to DNA replication by efficient extension from AraC-terminated primers. Here, we provide high-resolution structures of human Polη with AraC incorporated at the 3'-primer terminus. We show that Polη can accommodate AraC at different stages of the catalytic cycle, and that it can manipulate the conformation of the AraC sugar via specific hydrogen bonding and stacking interactions. Taken together, the structures provide a basis for the ability of Polη to extend DNA synthesis from AraC terminated primers.