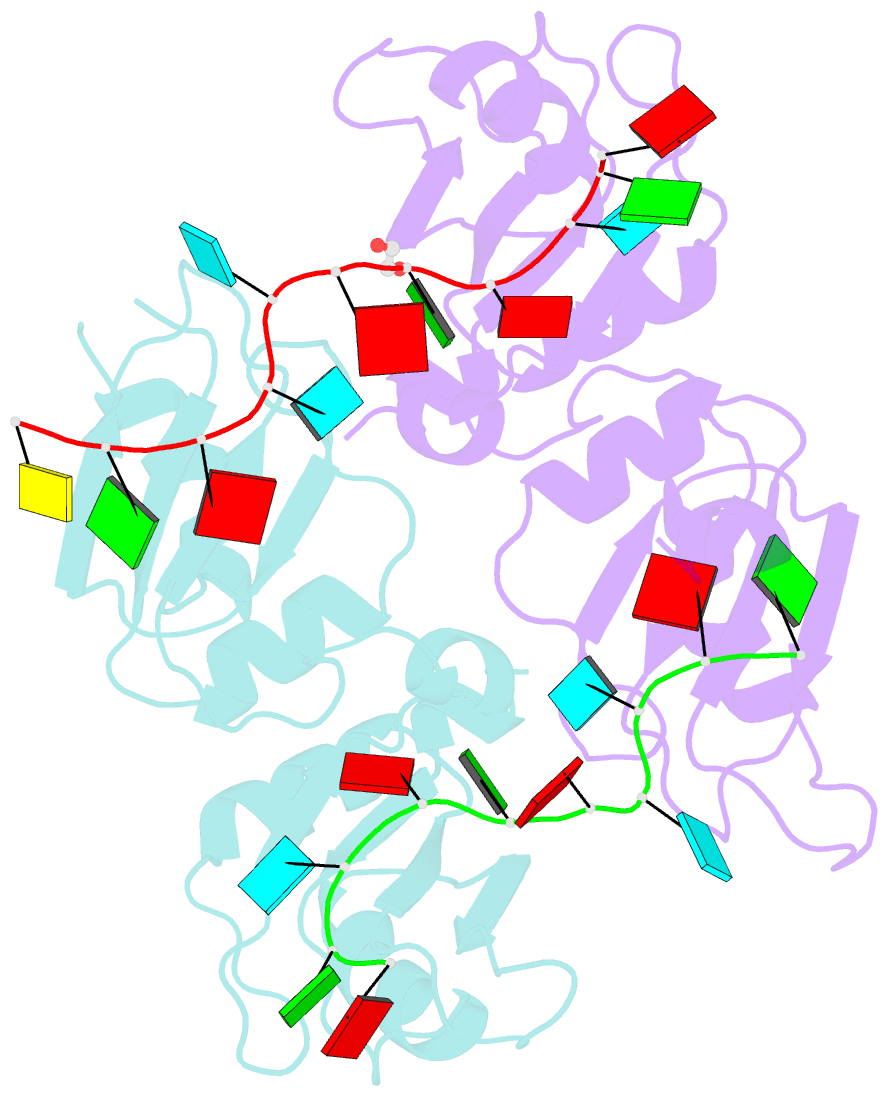
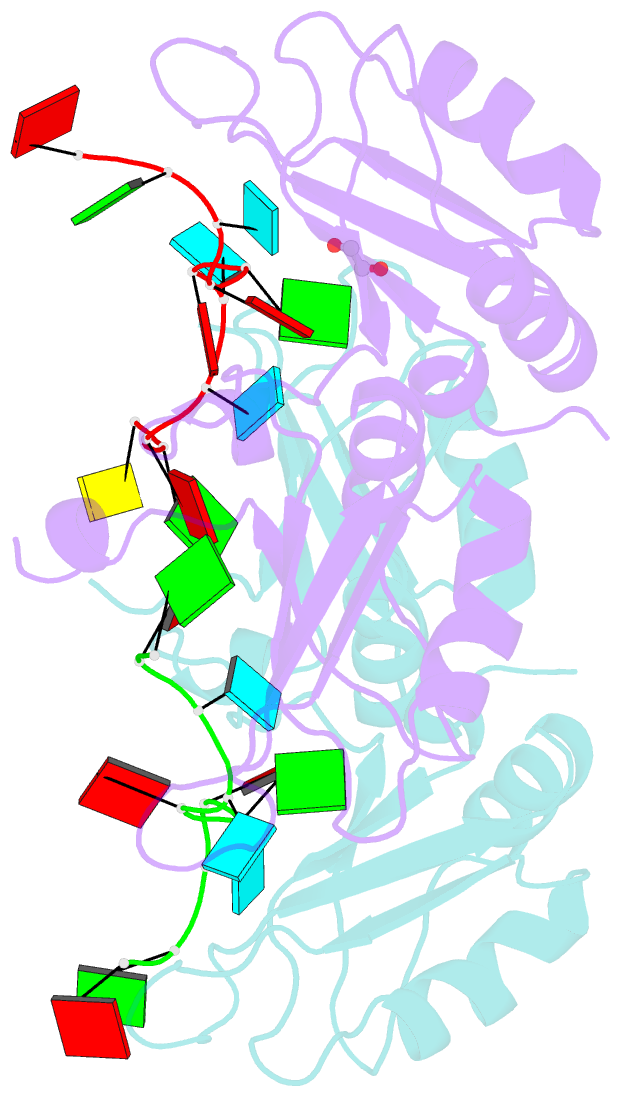
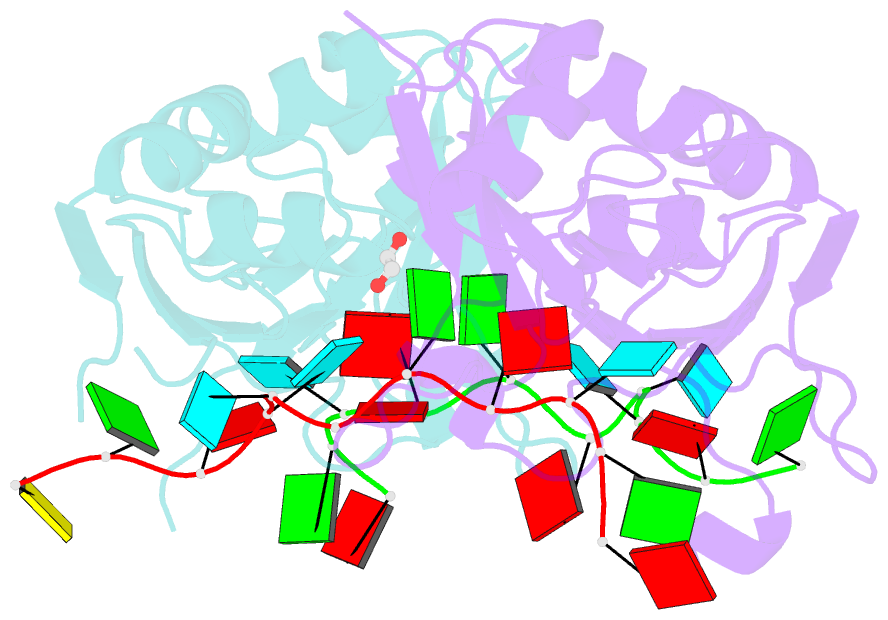
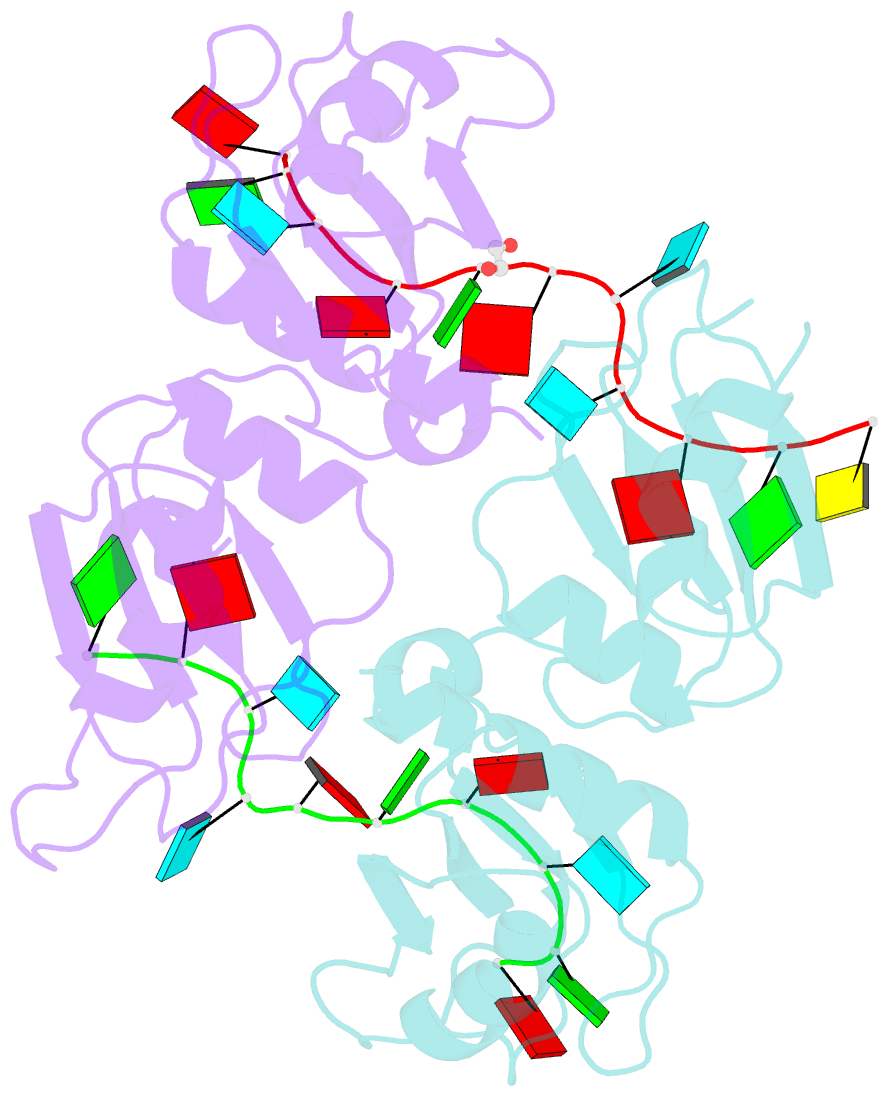
Summary information and primary citation
- PDB-id
- 6dcl; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.497 Å)
- Summary
- Crystal structure of up1 bound to pri-mirna-18a terminal loop
- Reference
- Kooshapur H, Choudhury NR, Simon B, Muhlbauer M, Jussupow A, Fernandez N, Jones AN, Dallmann A, Gabel F, Camilloni C, Michlewski G, Caceres JF, Sattler M (2018): "Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1." Nat Commun, 9, 2479. doi: 10.1038/s41467-018-04871-9.
- Abstract
- Post-transcriptional mechanisms play a predominant role in the control of microRNA (miRNA) production. Recognition of the terminal loop of precursor miRNAs by RNA-binding proteins (RBPs) influences their processing; however, the mechanistic basis for how levels of individual or subsets of miRNAs are regulated is mostly unexplored. We previously showed that hnRNP A1, an RBP implicated in many aspects of RNA processing, acts as an auxiliary factor that promotes the Microprocessor-mediated processing of pri-mir-18a. Here, by using an integrative structural biology approach, we show that hnRNP A1 forms a 1:1 complex with pri-mir-18a where both RNA recognition motifs (RRMs) bind to cognate RNA sequence motifs in the terminal loop of pri-mir-18a. Terminal loop binding induces an allosteric destabilization of base-pairing in the pri-mir-18a stem that promotes its downstream processing. Our results highlight terminal loop RNA recognition by RBPs as a potential general principle of miRNA biogenesis and regulation.