Summary information and primary citation
- PDB-id
- 6dnh; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.4 Å)
- Summary
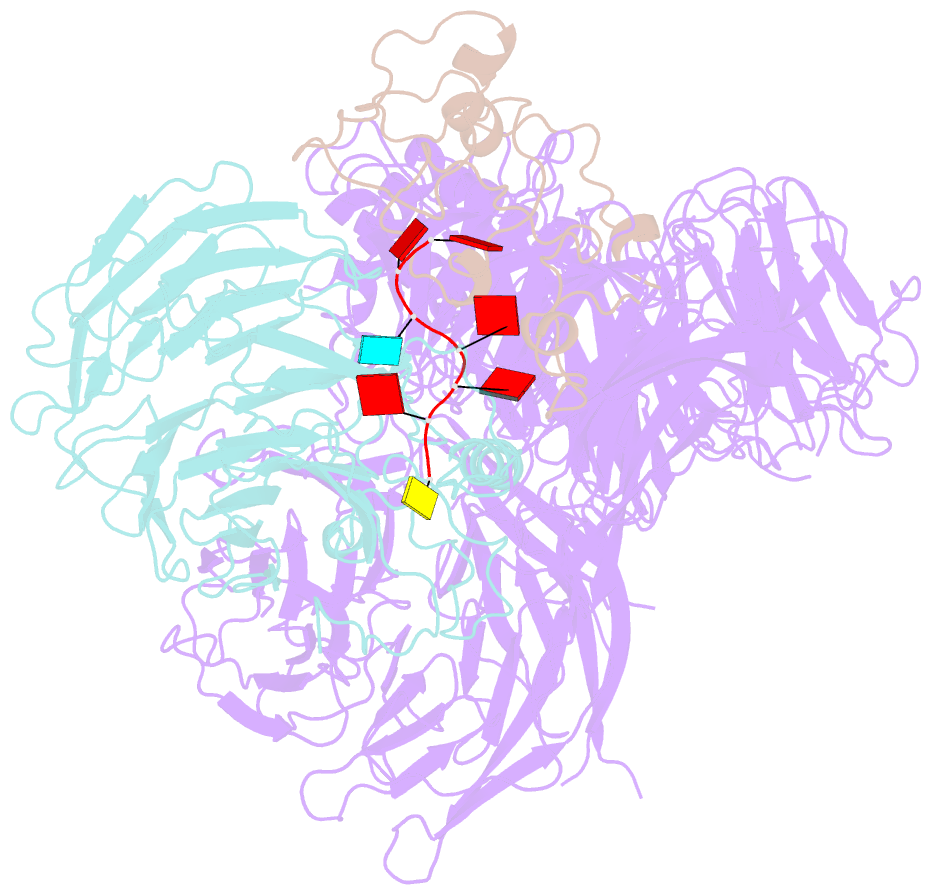
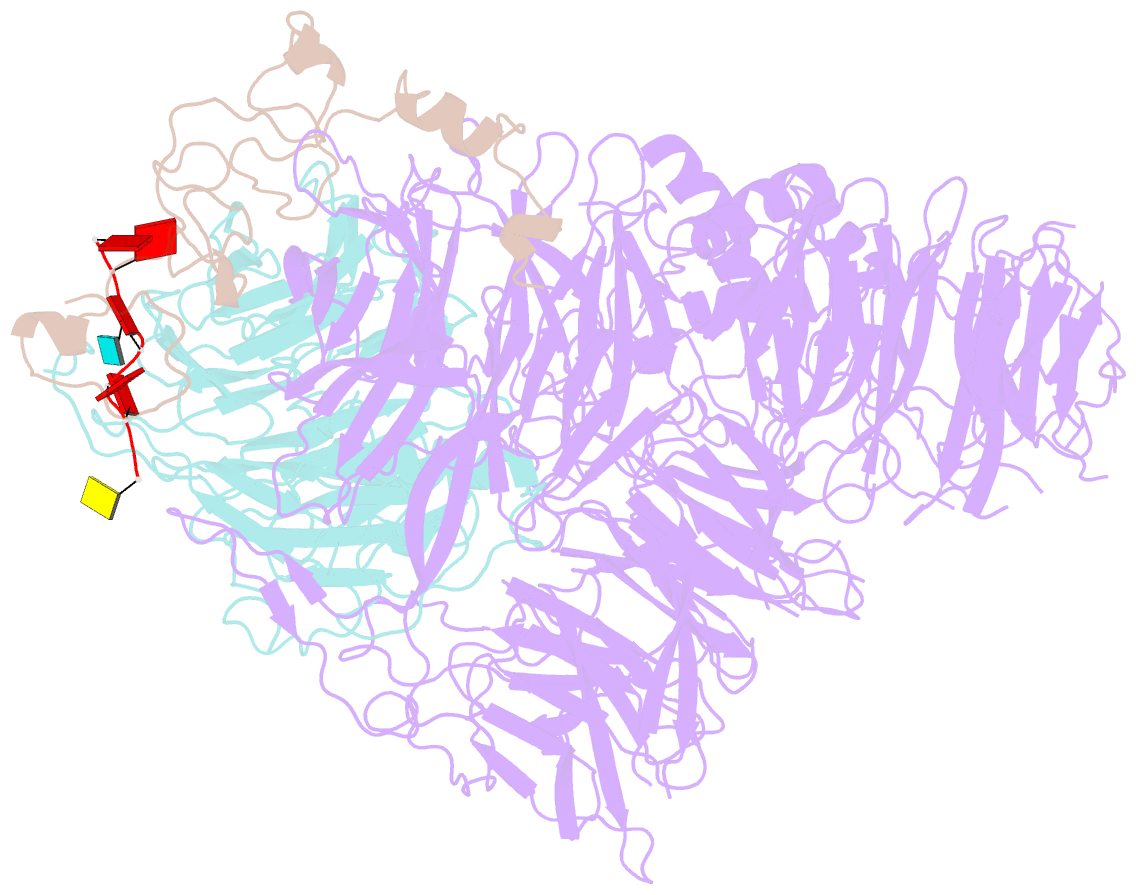
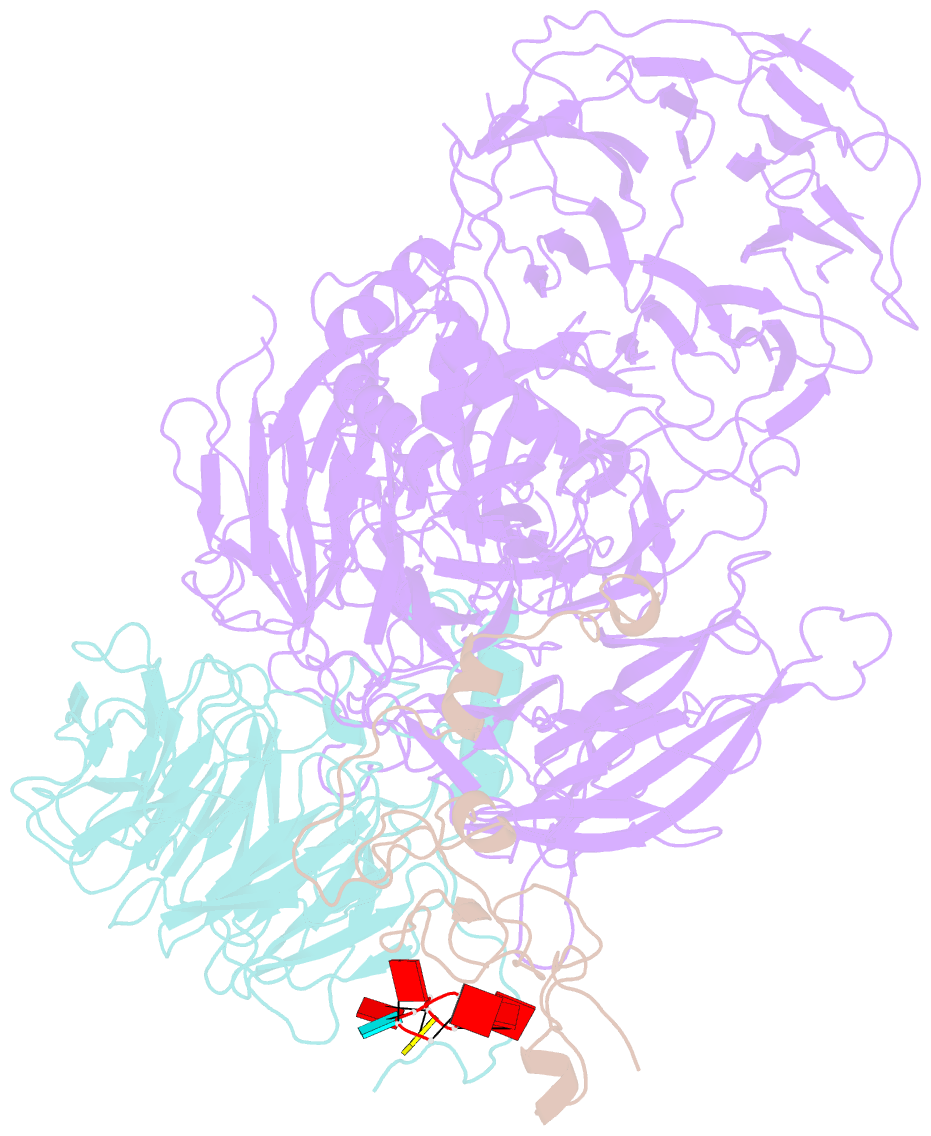
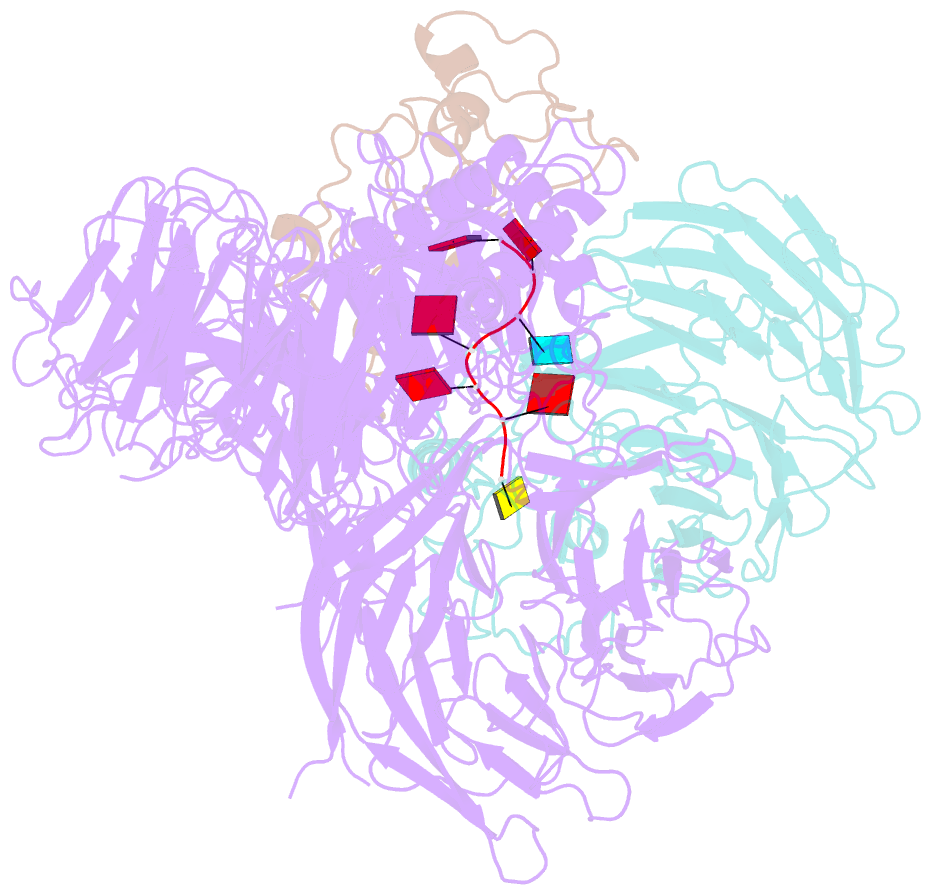
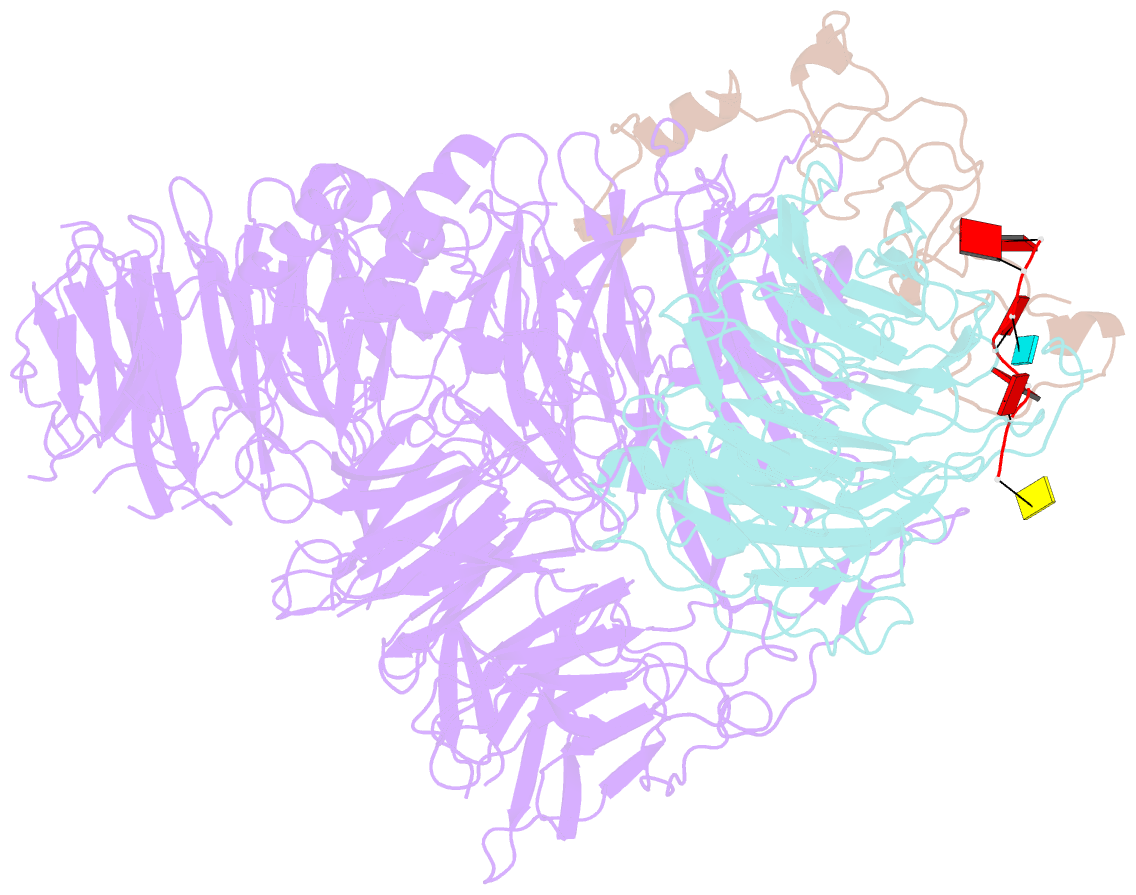
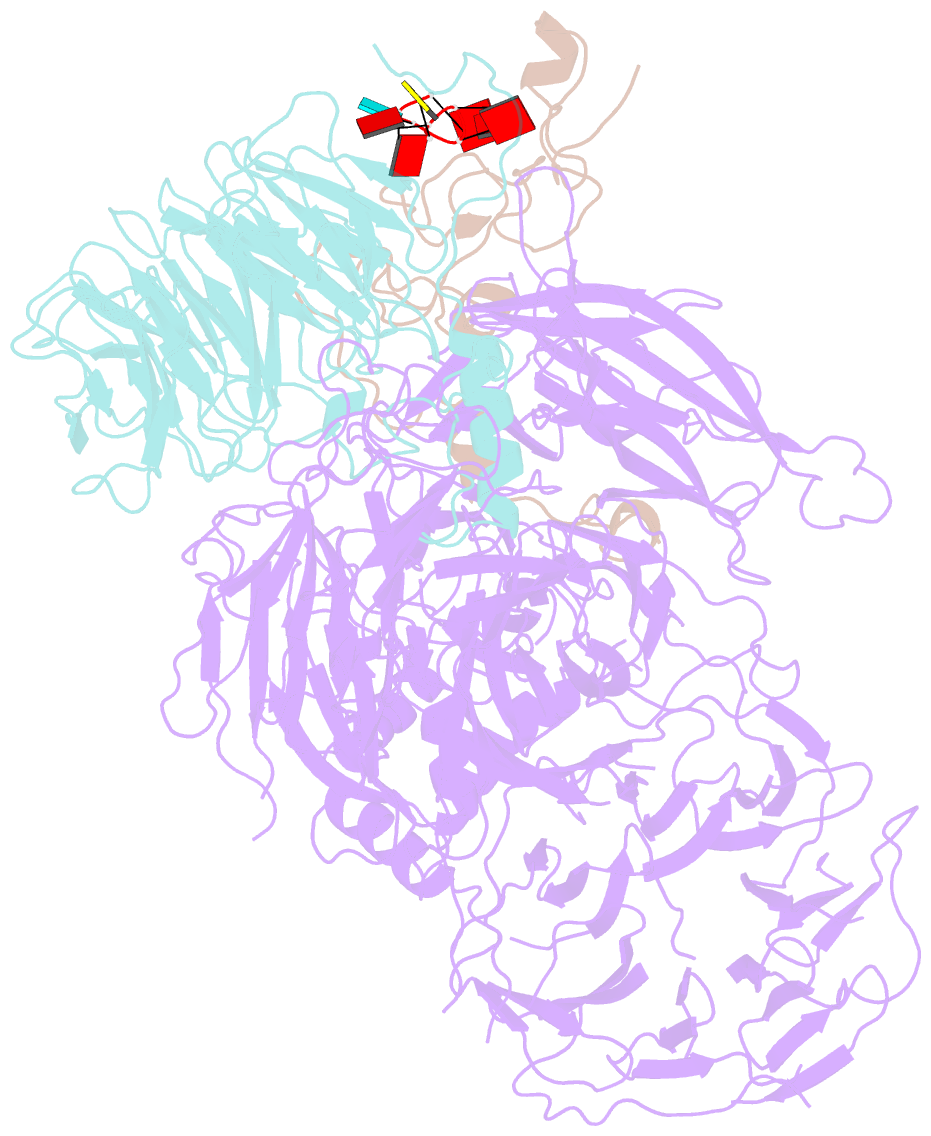
- cryo-EM structure of human cpsf-160-wdr33-cpsf-30-pas RNA complex at 3.4 Å resolution
- Reference
- Sun Y, Zhang Y, Hamilton K, Manley JL, Shi Y, Walz T, Tong L (2018): "Molecular basis for the recognition of the human AAUAAA polyadenylation signal." Proc. Natl. Acad. Sci. U.S.A., 115, E1419-E1428. doi: 10.1073/pnas.1718723115.
- Abstract
- Nearly all eukaryotic messenger RNA precursors must undergo cleavage and polyadenylation at their 3'-end for maturation. A crucial step in this process is the recognition of the AAUAAA polyadenylation signal (PAS), and the molecular mechanism of this recognition has been a long-standing problem. Here, we report the cryo-electron microscopy structure of a quaternary complex of human CPSF-160, WDR33, CPSF-30, and an AAUAAA RNA at 3.4-Å resolution. Strikingly, the AAUAAA PAS assumes an unusual conformation that allows this short motif to be bound directly by both CPSF-30 and WDR33. The A1 and A2 bases are recognized specifically by zinc finger 2 (ZF2) of CPSF-30 and the A4 and A5 bases by ZF3. Interestingly, the U3 and A6 bases form an intramolecular Hoogsteen base pair and directly contact WDR33. CPSF-160 functions as an essential scaffold and preorganizes CPSF-30 and WDR33 for high-affinity binding to AAUAAA. Our findings provide an elegant molecular explanation for how PAS sequences are recognized for mRNA 3'-end formation.