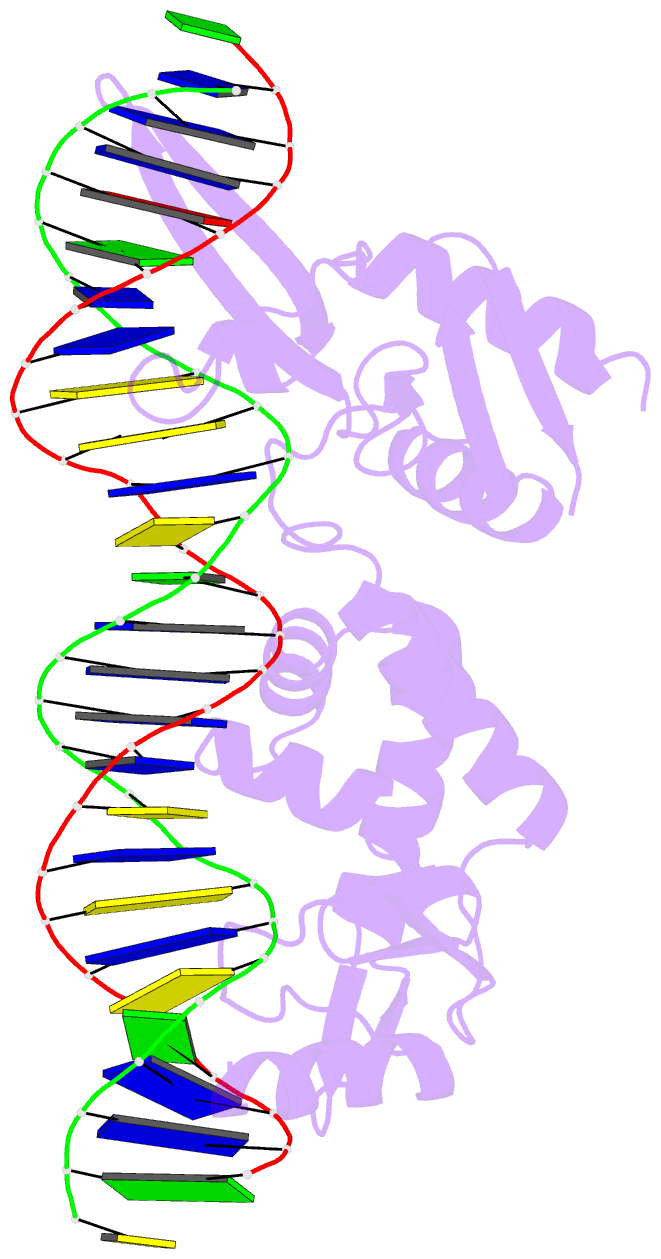
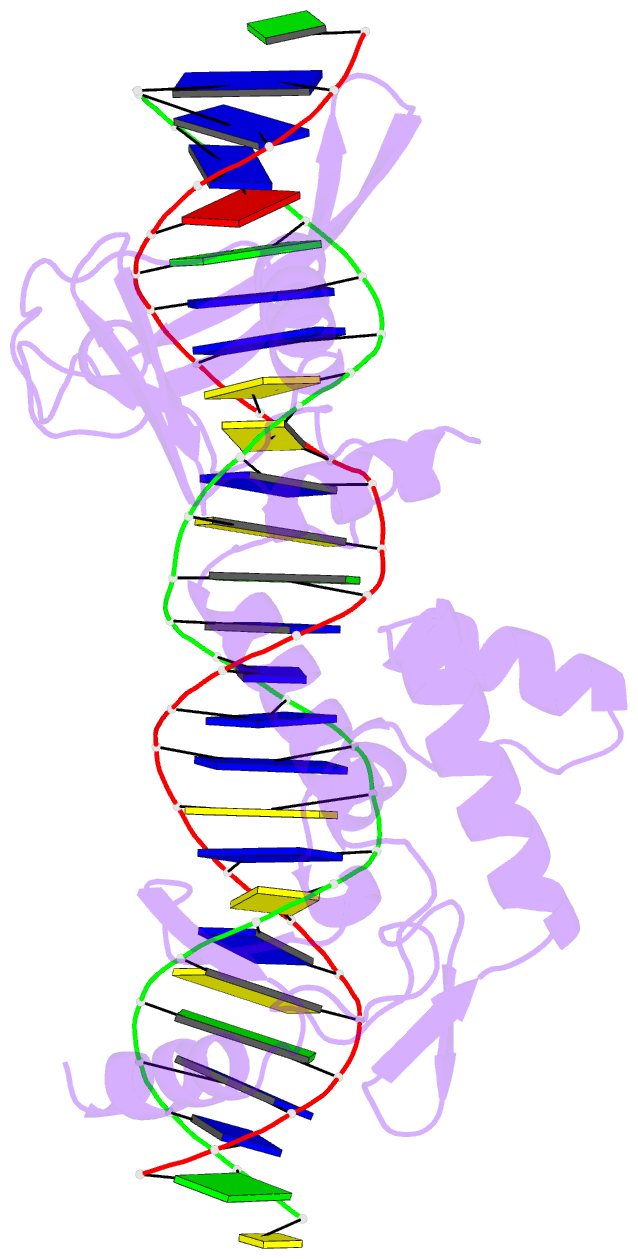
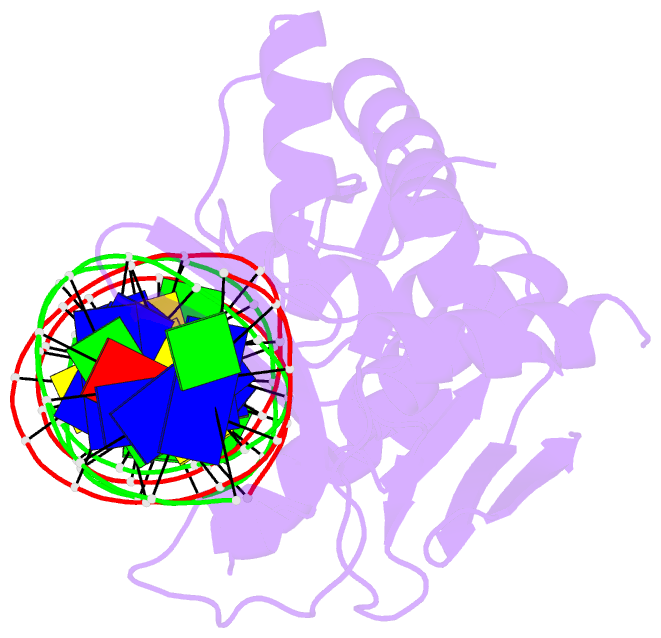
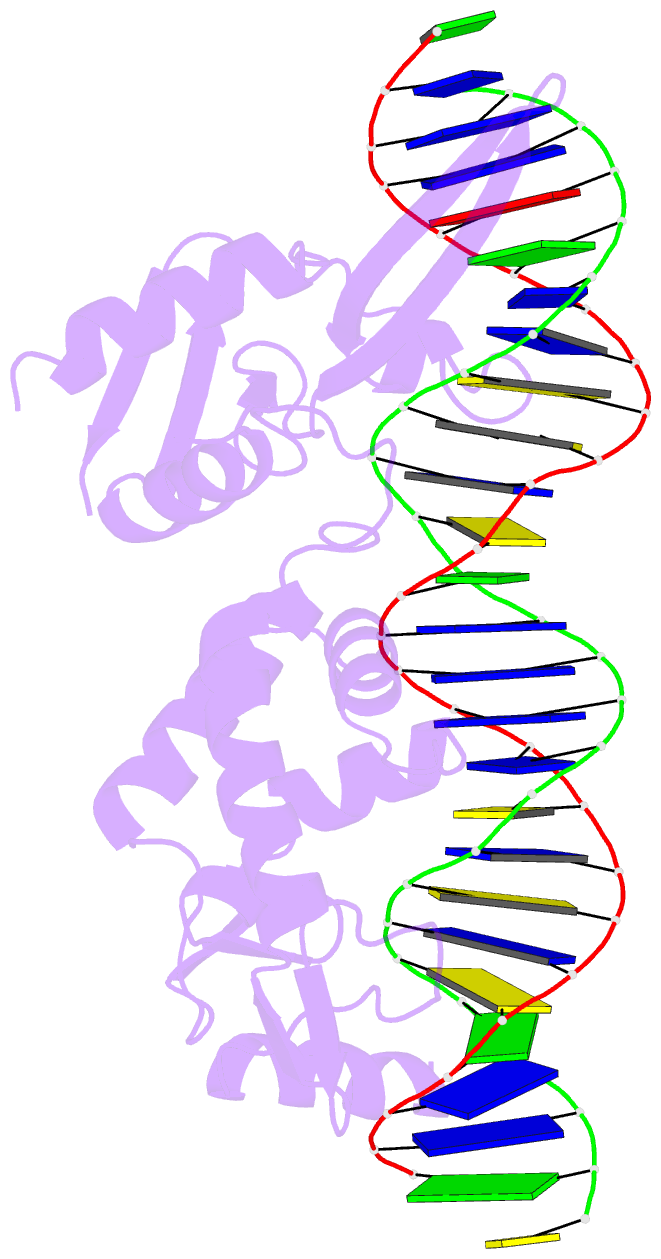
Summary information and primary citation
- PDB-id
- 6dnw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.849 Å)
- Summary
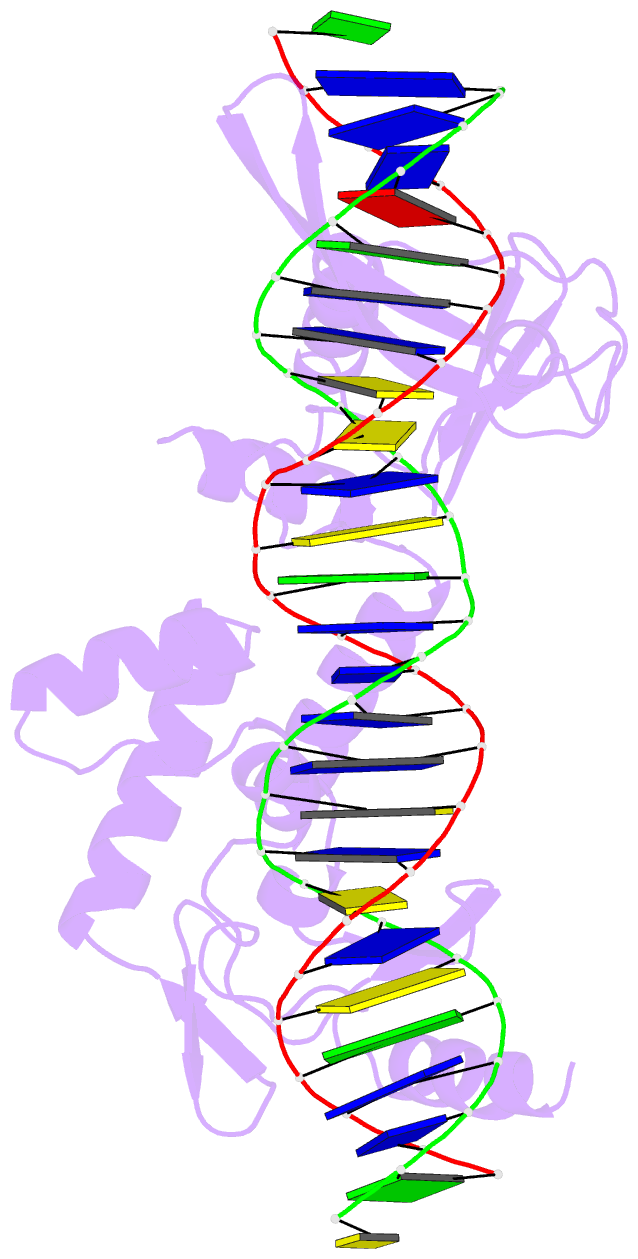
- Sequence requirements of the listeria innocua prophage attp site
- Reference
- Li H, Sharp R, Rutherford K, Gupta K, Van Duyne GD (2018): "Serine Integrase attP Binding and Specificity." J. Mol. Biol., 430, 4401-4418. doi: 10.1016/j.jmb.2018.09.007.
- Abstract
- Serine integrases catalyze the site-specific insertion of viral DNA into a host's genome. The minimal requirements and irreversible nature of this integration reaction have led to the use of serine integrases in applications ranging from bacterial memory storage devices to gene therapy. Our understanding of how the integrase proteins recognize the viral (attP) and host (attB) attachment sites is limited, with structural data available for only a Listeria integrase C-terminal domain (CTD) bound to an attP half-site. Here we report quantitative binding and saturation mutagenesis analyses for the Listeria innocua prophage attP site and a new 2.8-Å crystal structure of the CTD•attP half site. We find that Int binds with high affinity to attP (6.9 nM), but the Int CTD binds to attP half-sites with only 7- to 10-fold lower affinity, supporting the idea that free energy is expended to open an Int dimer for attP binding. Despite the 50-bp Int-attP interaction surface, only 20 residues are sensitive to mutagenesis, and of these, only 6 require a specific residue for efficient Int binding and integration activity. One of the integrase DNA-binding domains, the recombinase domain, appears to be primarily non-specific. Several substitutions result in an improved attP site, indicating that higher-efficiency attachment sites can be obtained through site engineering. These findings advance our understanding of serine integrase function and provide important data for efforts towards engineering this family of enzymes for a variety of biotechnology applications.