Summary information and primary citation
- PDB-id
- 6dt1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-DNA
- Method
- X-ray (2.75 Å)
- Summary
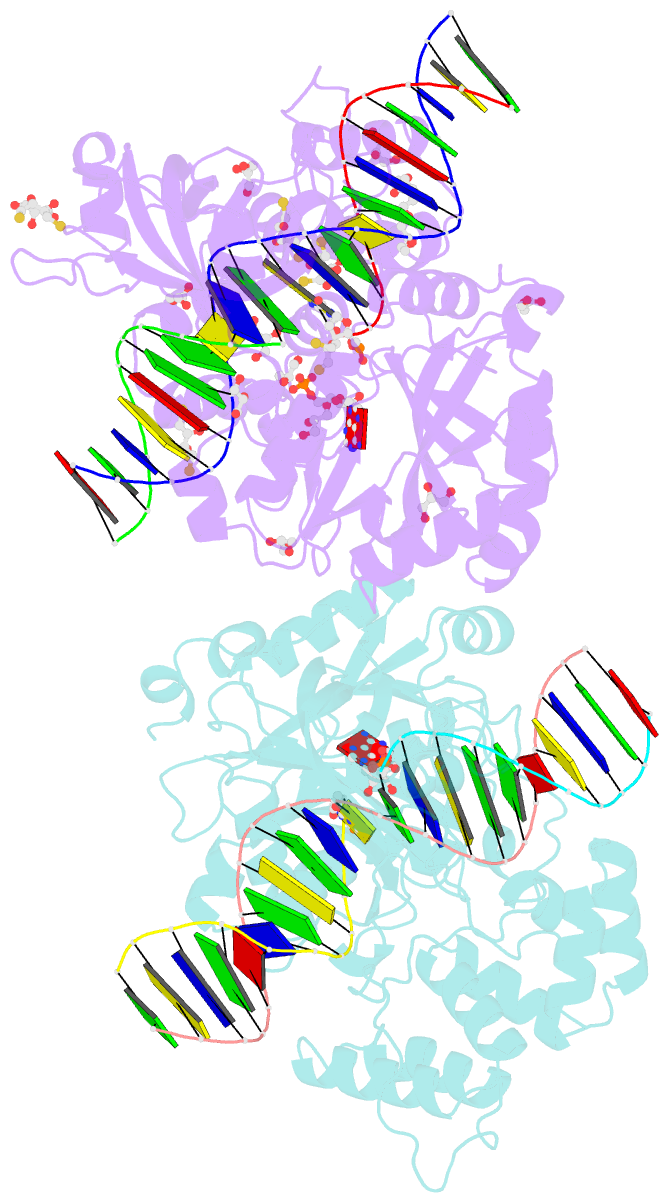
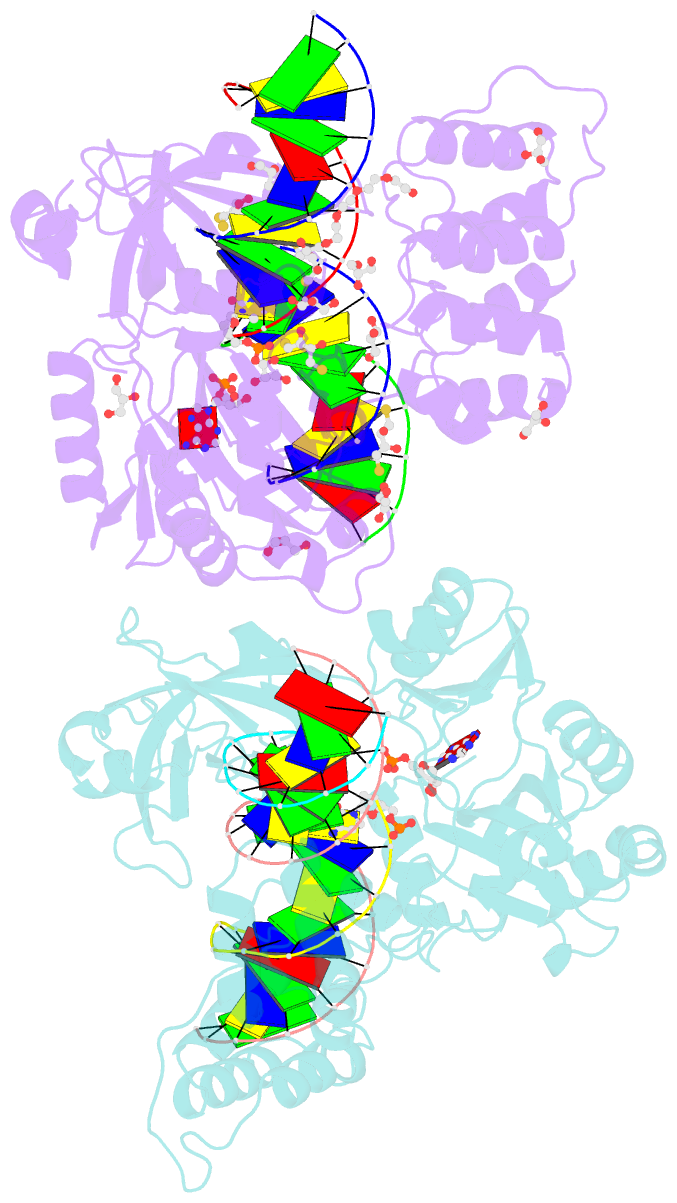
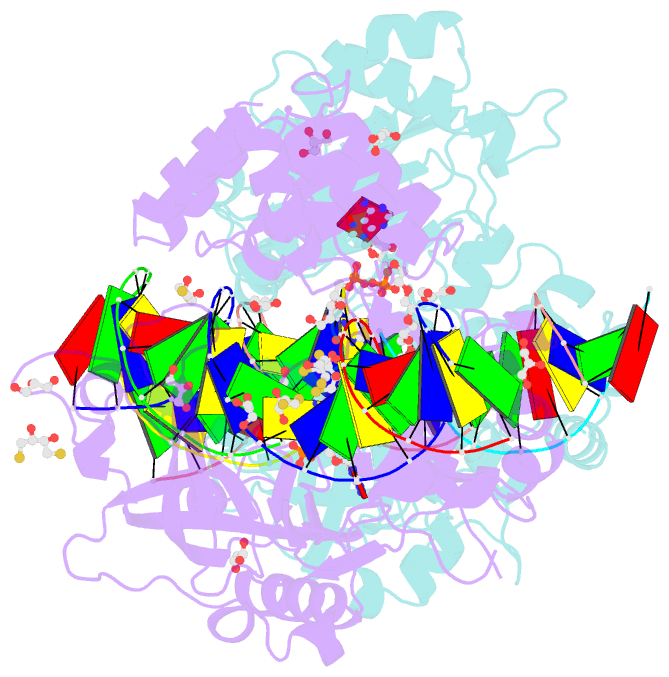
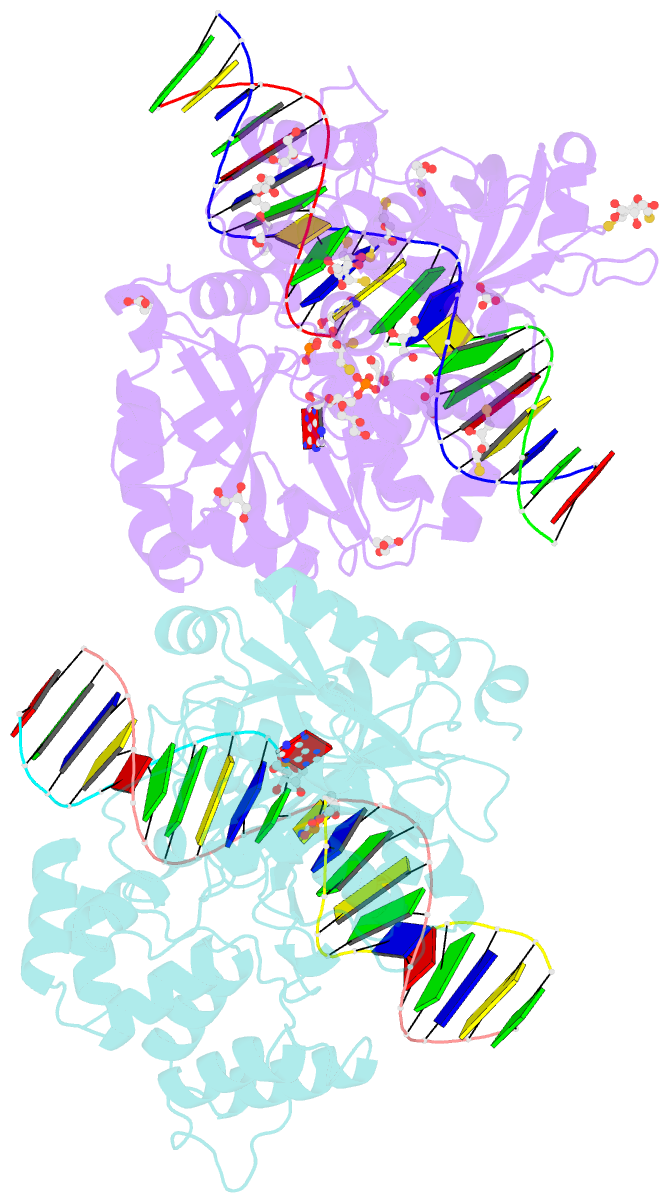
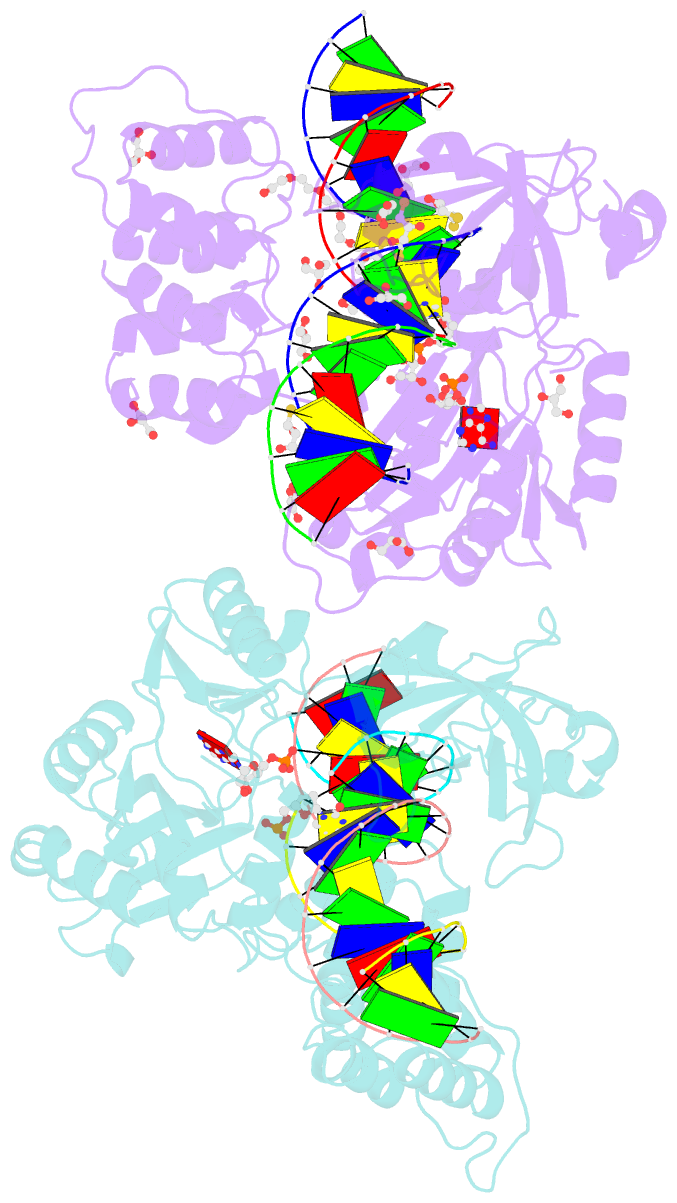
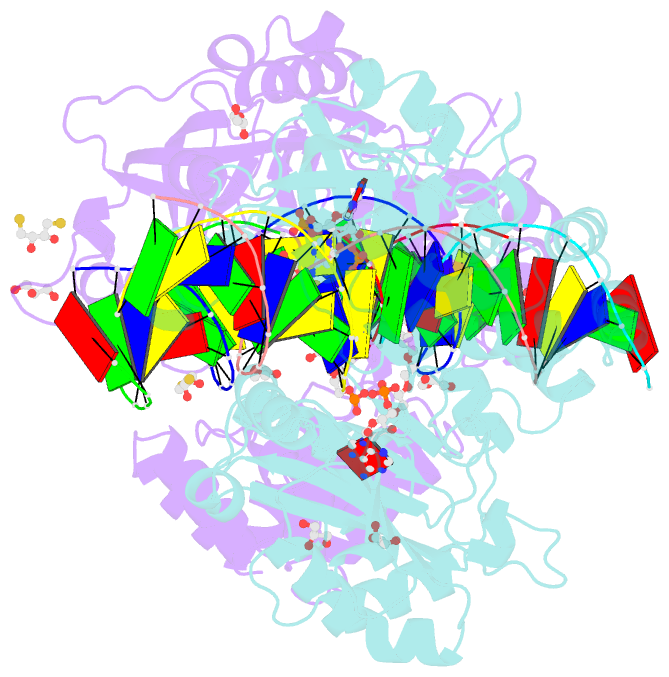
- Crystal structure of the ligase from bacteriophage t4 complexed with DNA intermediate
- Reference
- Shi K, Bohl TE, Park J, Zasada A, Malik S, Banerjee S, Tran V, Li N, Yin Z, Kurniawan F, Orellana K, Aihara H (2018): "T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction." Nucleic Acids Res., 46, 10474-10488. doi: 10.1093/nar/gky776.
- Abstract
- DNA ligases play essential roles in DNA replication and repair. Bacteriophage T4 DNA ligase is the first ATP-dependent ligase enzyme to be discovered and is widely used in molecular biology, but its structure remained unknown. Our crystal structure of T4 DNA ligase bound to DNA shows a compact α-helical DNA-binding domain (DBD), nucleotidyl-transferase (NTase) domain, and OB-fold domain, which together fully encircle DNA. The DBD of T4 DNA ligase exhibits remarkable structural homology to the core DNA-binding helices of the larger DBDs from eukaryotic and archaeal DNA ligases, but it lacks additional structural components required for protein interactions. T4 DNA ligase instead has a flexible loop insertion within the NTase domain, which binds tightly to the T4 sliding clamp gp45 in a novel α-helical PIP-box conformation. Thus, T4 DNA ligase represents a prototype of the larger eukaryotic and archaeal DNA ligases, with a uniquely evolved mode of protein interaction that may be important for efficient DNA replication.