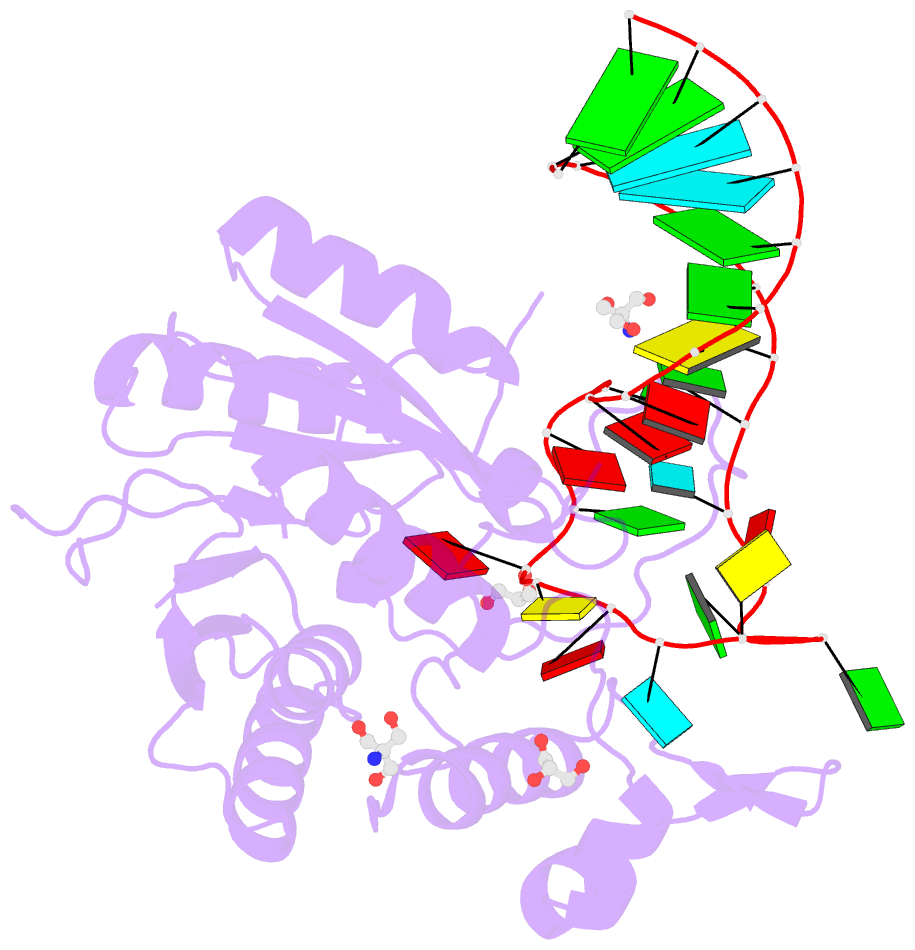
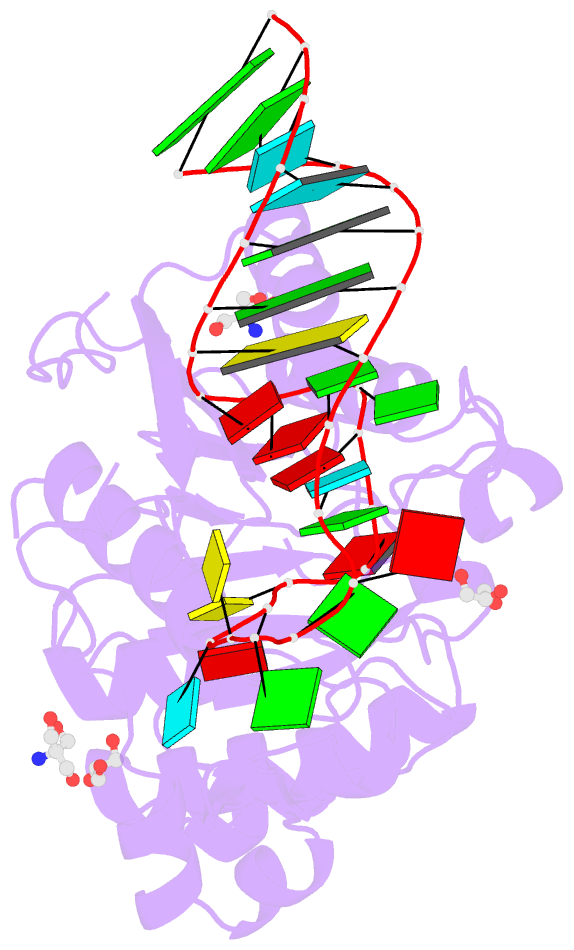
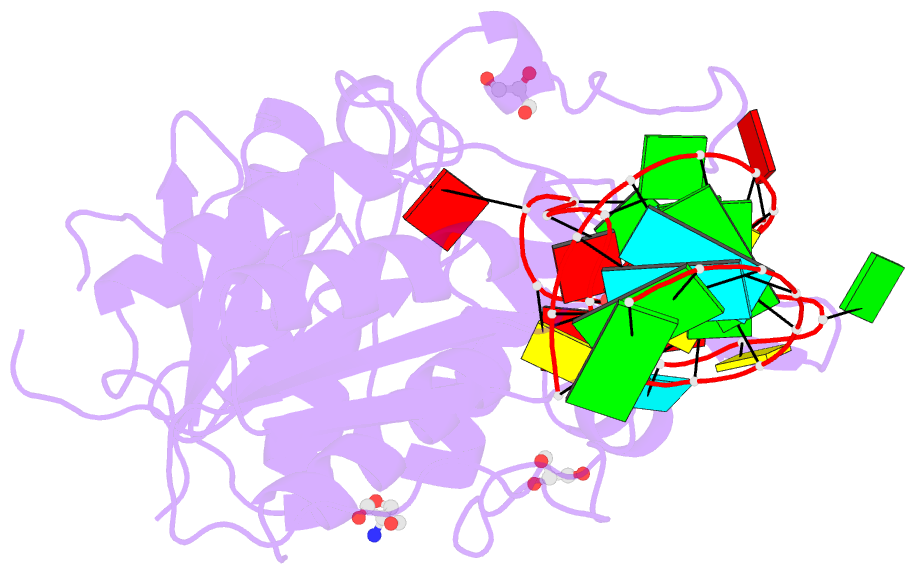
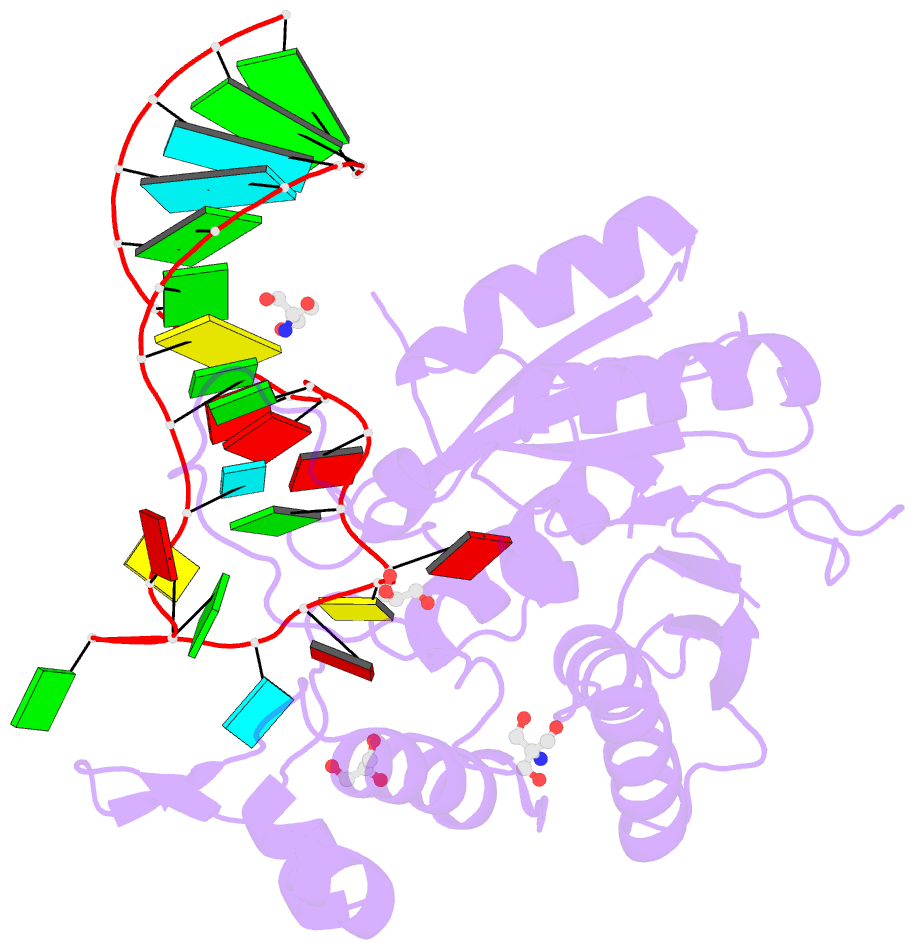
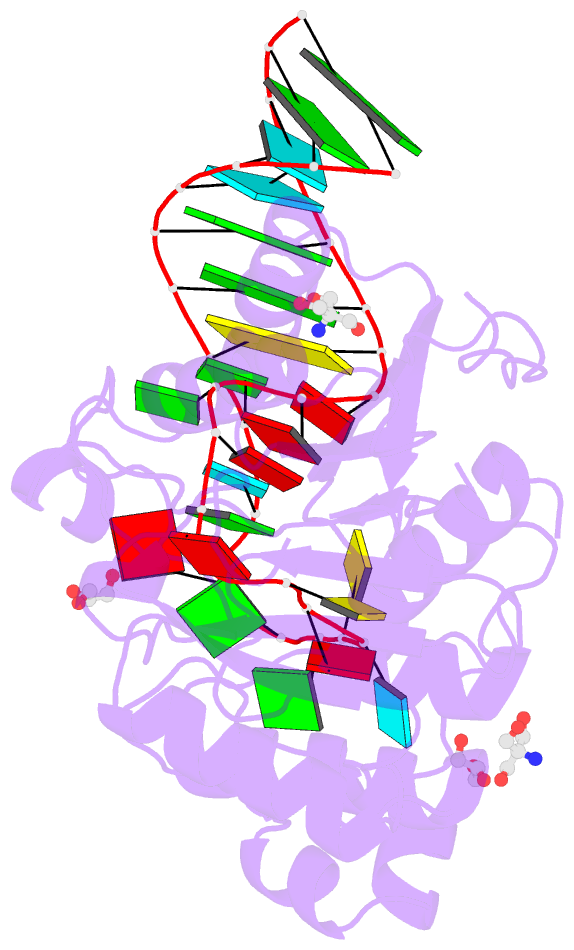
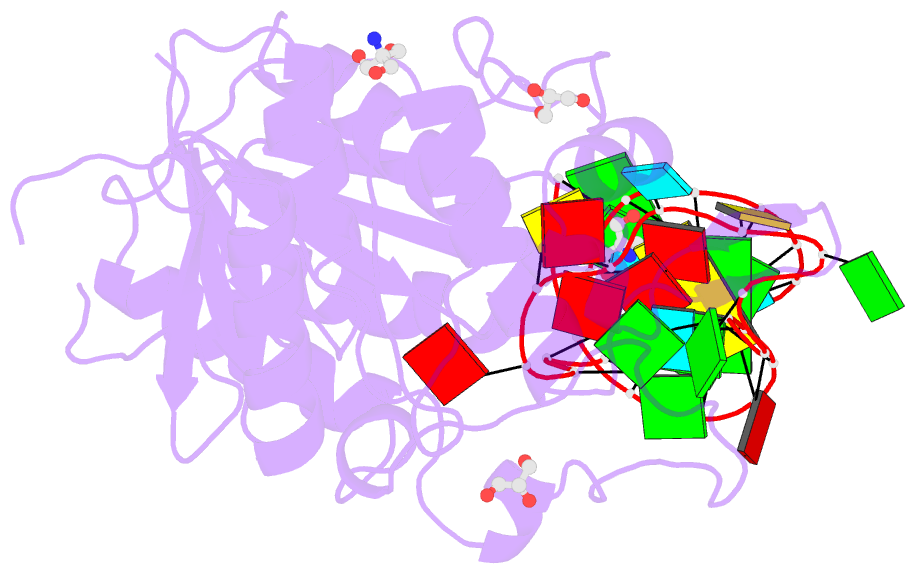
Summary information and primary citation
- PDB-id
- 6du4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (1.7 Å)
- Summary
- Crystal structure of hmettl16 catalytic domain in complex with mat2a 3'utr hairpin 1
- Reference
- Doxtader KA, Wang P, Scarborough AM, Seo D, Conrad NK, Nam Y (2018): "Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor." Mol. Cell, 71, 1001-1011.e4. doi: 10.1016/j.molcel.2018.07.025.
- Abstract
- S-adenosylmethionine (SAM) is an essential metabolite that acts as a cofactor for most methylation events in the cell. The N6-methyladenosine (m6A) methyltransferase METTL16 controls SAM homeostasis by regulating the abundance of SAM synthetase MAT2A mRNA in response to changing intracellular SAM levels. Here we present crystal structures of METTL16 in complex with MAT2A RNA hairpins to uncover critical molecular mechanisms underlying the regulated activity of METTL16. The METTL16-RNA complex structures reveal atomic details of RNA substrates that drive productive methylation by METTL16. In addition, we identify a polypeptide loop in METTL16 near the SAM binding site with an autoregulatory role. We show that mutations that enhance or repress METTL16 activity in vitro correlate with changes in MAT2A mRNA levels in cells. Thus, we demonstrate the structural basis for the specific activity of METTL16 and further suggest the molecular mechanisms by which METTL16 efficiency is tuned to regulate SAM homeostasis.