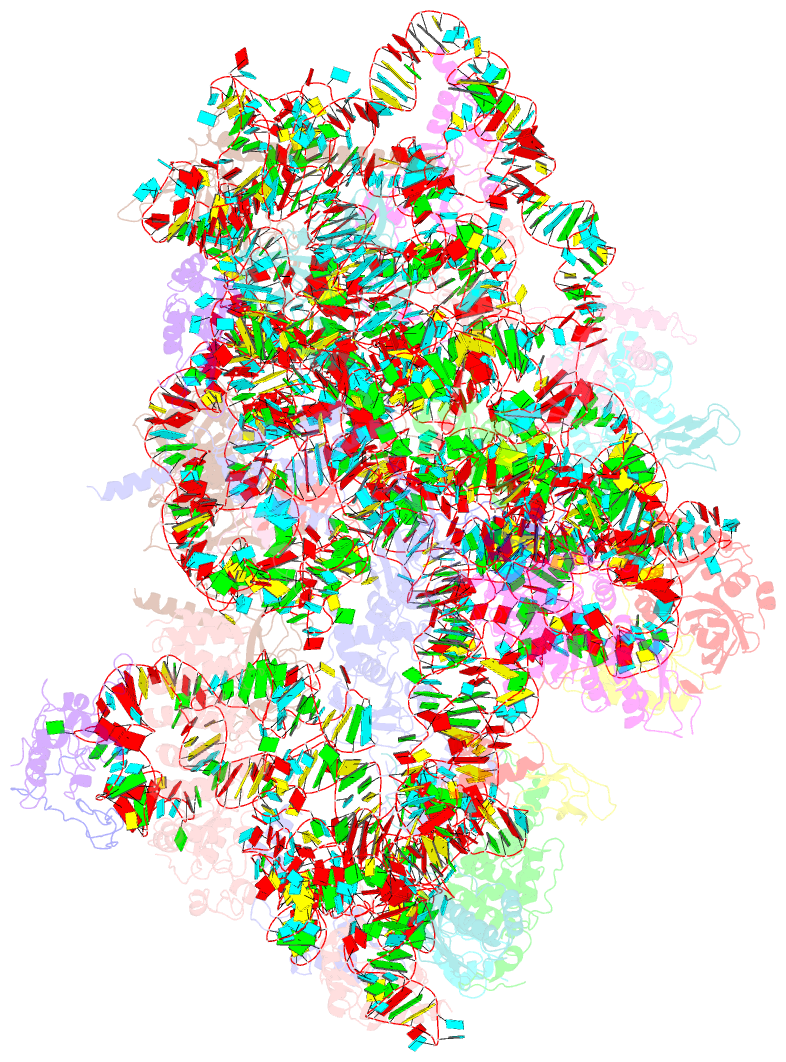
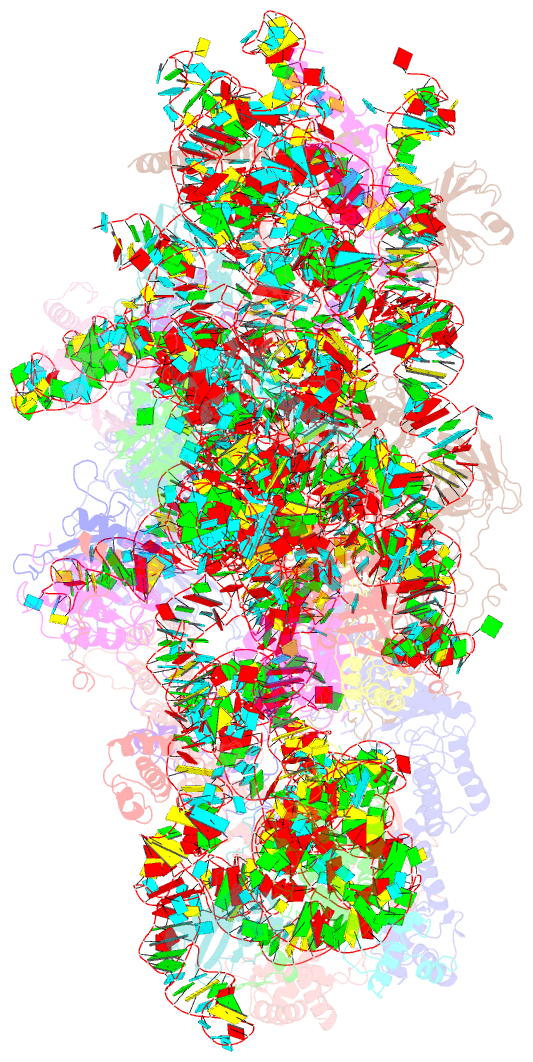
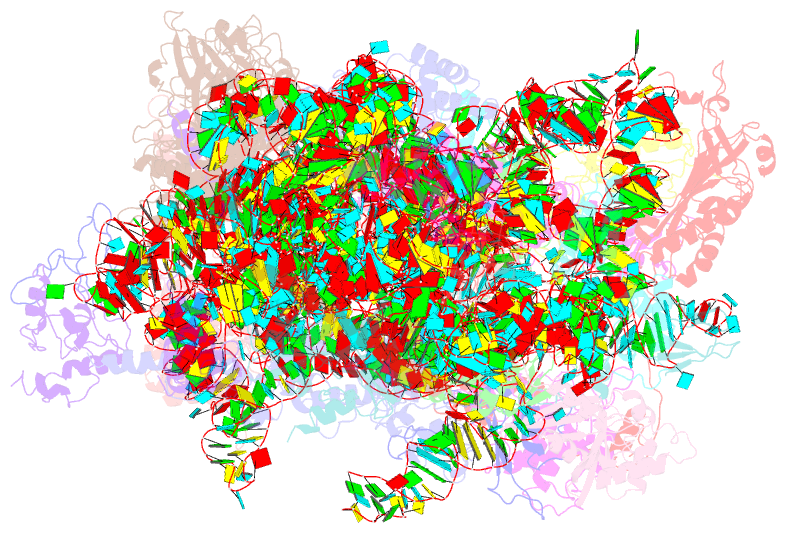
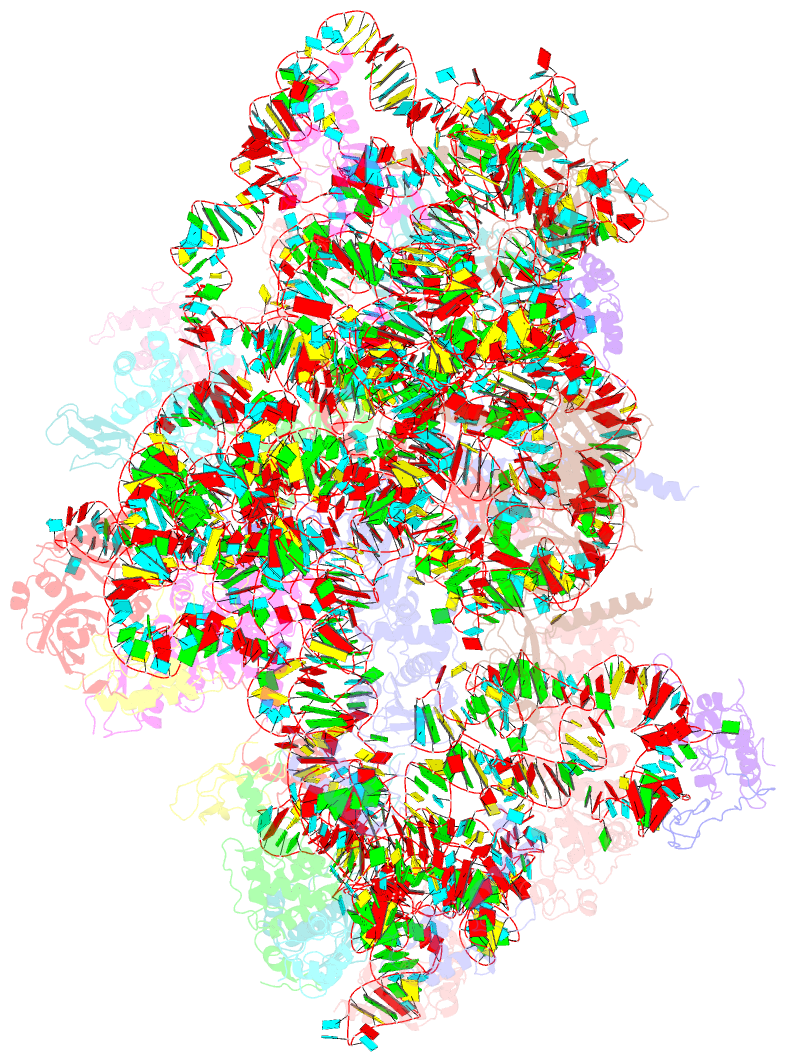
Summary information and primary citation
- PDB-id
- 6eml; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (3.6 Å)
- Summary
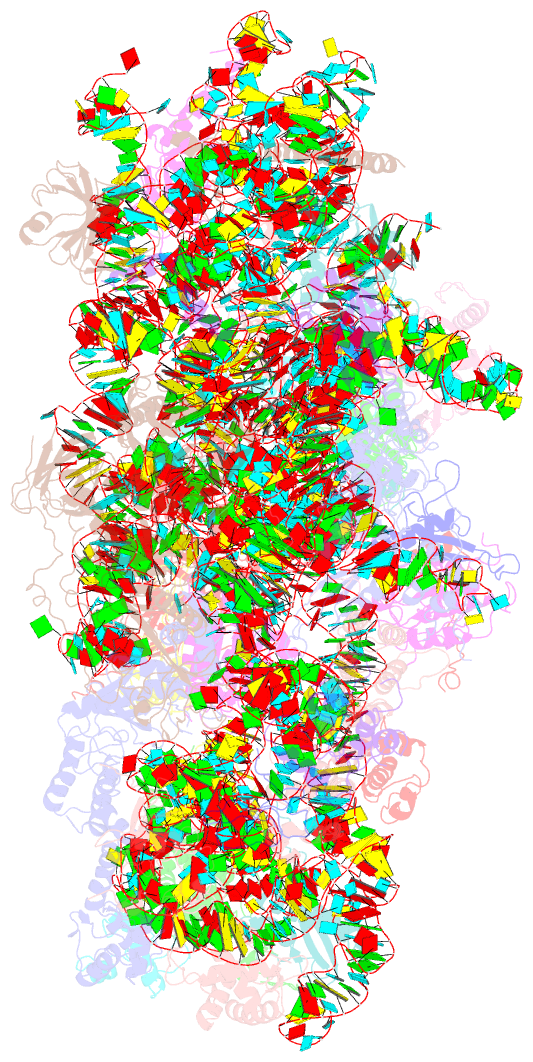
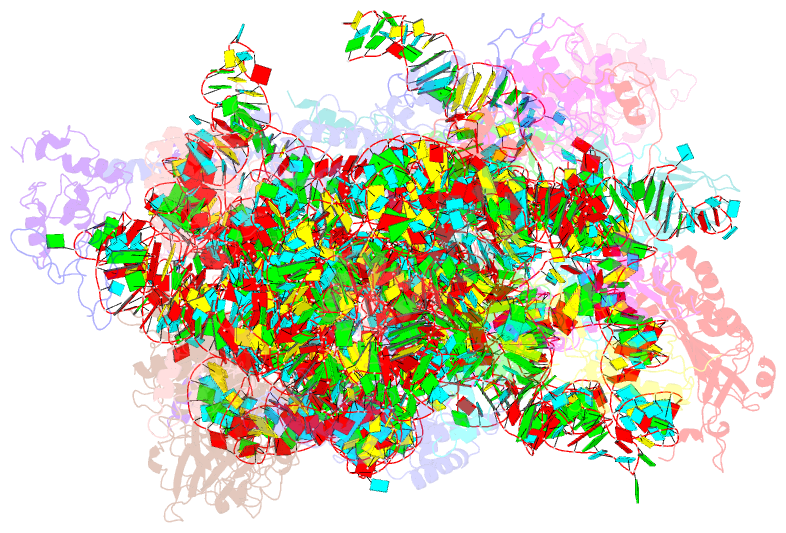
- cryo-EM structure of a late pre-40s ribosomal subunit from saccharomyces cerevisiae
- Reference
- Heuer A, Thomson E, Schmidt C, Berninghausen O, Becker T, Hurt E, Beckmann R (2017): "Cryo-EM structure of a late pre-40S ribosomal subunit fromSaccharomyces cerevisiae." Elife, 6. doi: 10.7554/eLife.30189.
- Abstract
- Mechanistic understanding of eukaryotic ribosome formation requires a detailed structural knowledge of the numerous assembly intermediates, generated along a complex pathway. Here, we present the structure of a late pre-40S particle at 3.6 Å resolution, revealing in molecular detail how assembly factors regulate the timely folding of pre-18S rRNA. The structure shows that, rather than sterically blocking 40S translational active sites, the associated assembly factors Tsr1, Enp1, Rio2 and Pno1 collectively preclude their final maturation, thereby preventing untimely tRNA and mRNA binding and error prone translation. Moreover, the structure explains how Pno1 coordinates the 3'end cleavage of the 18S rRNA by Nob1 and how the late factor's removal in the cytoplasm ensures the structural integrity of the maturing 40S subunit.