Summary information and primary citation
- PDB-id
- 6en8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (3.29 Å)
- Summary
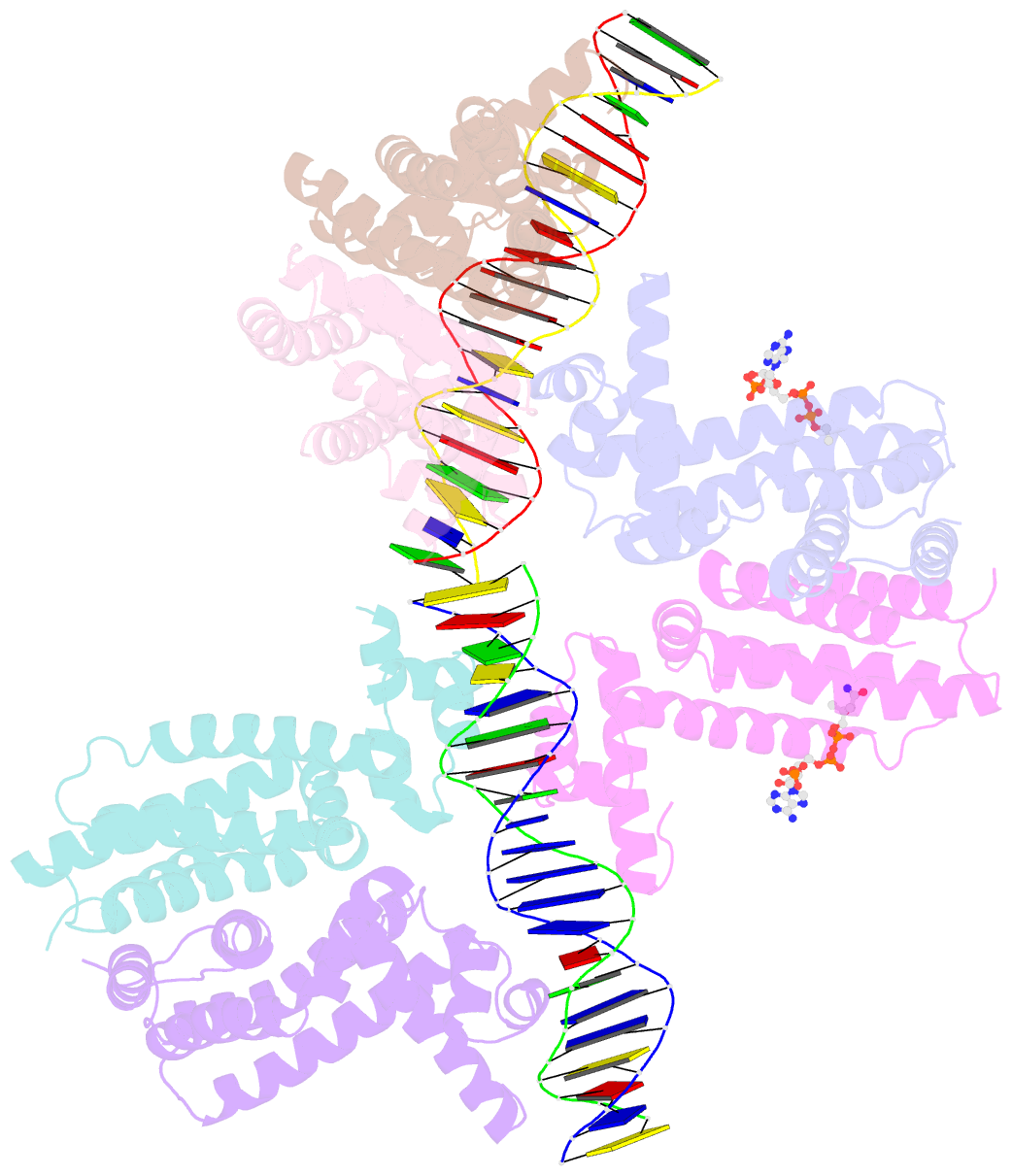
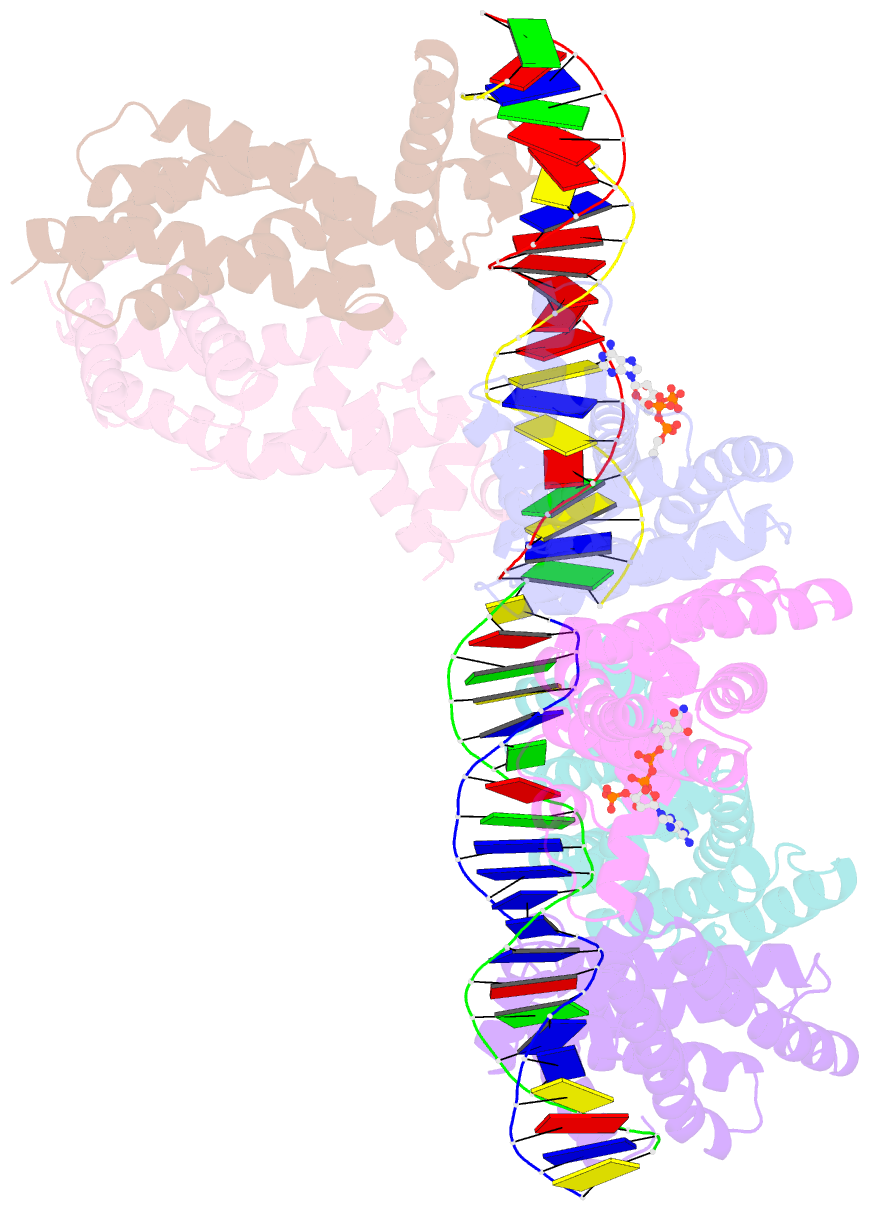
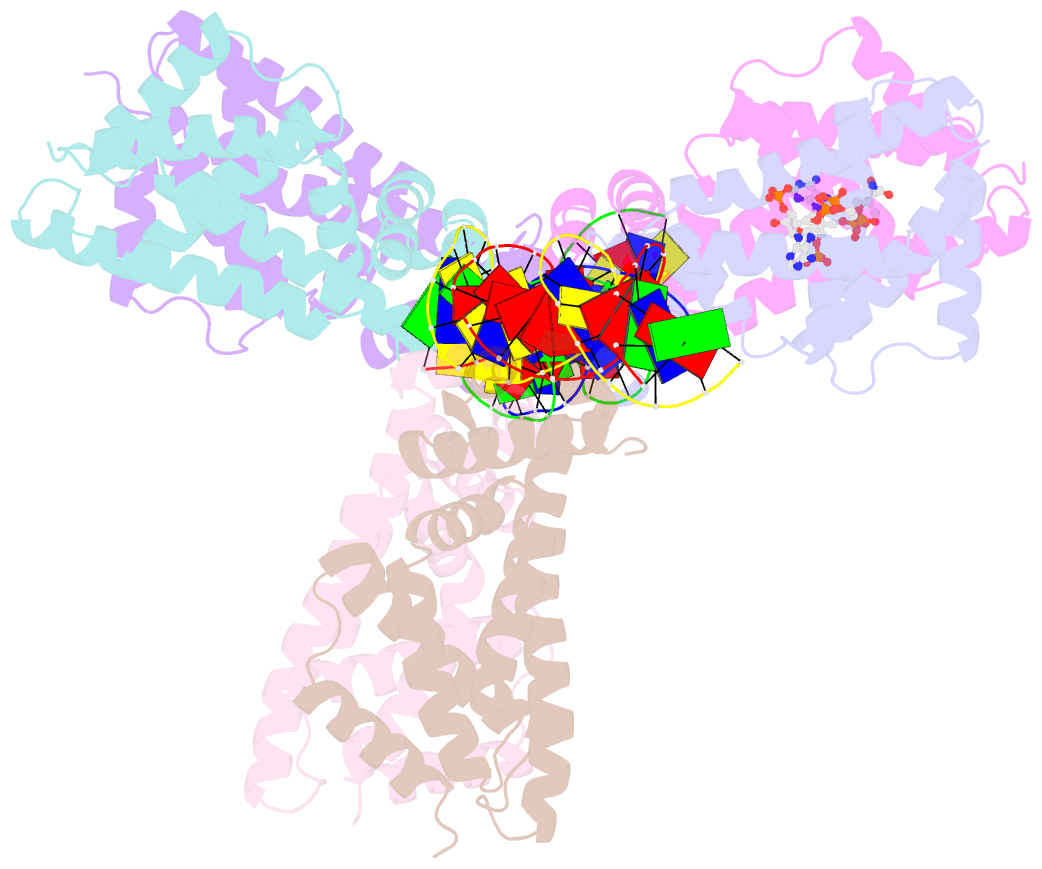
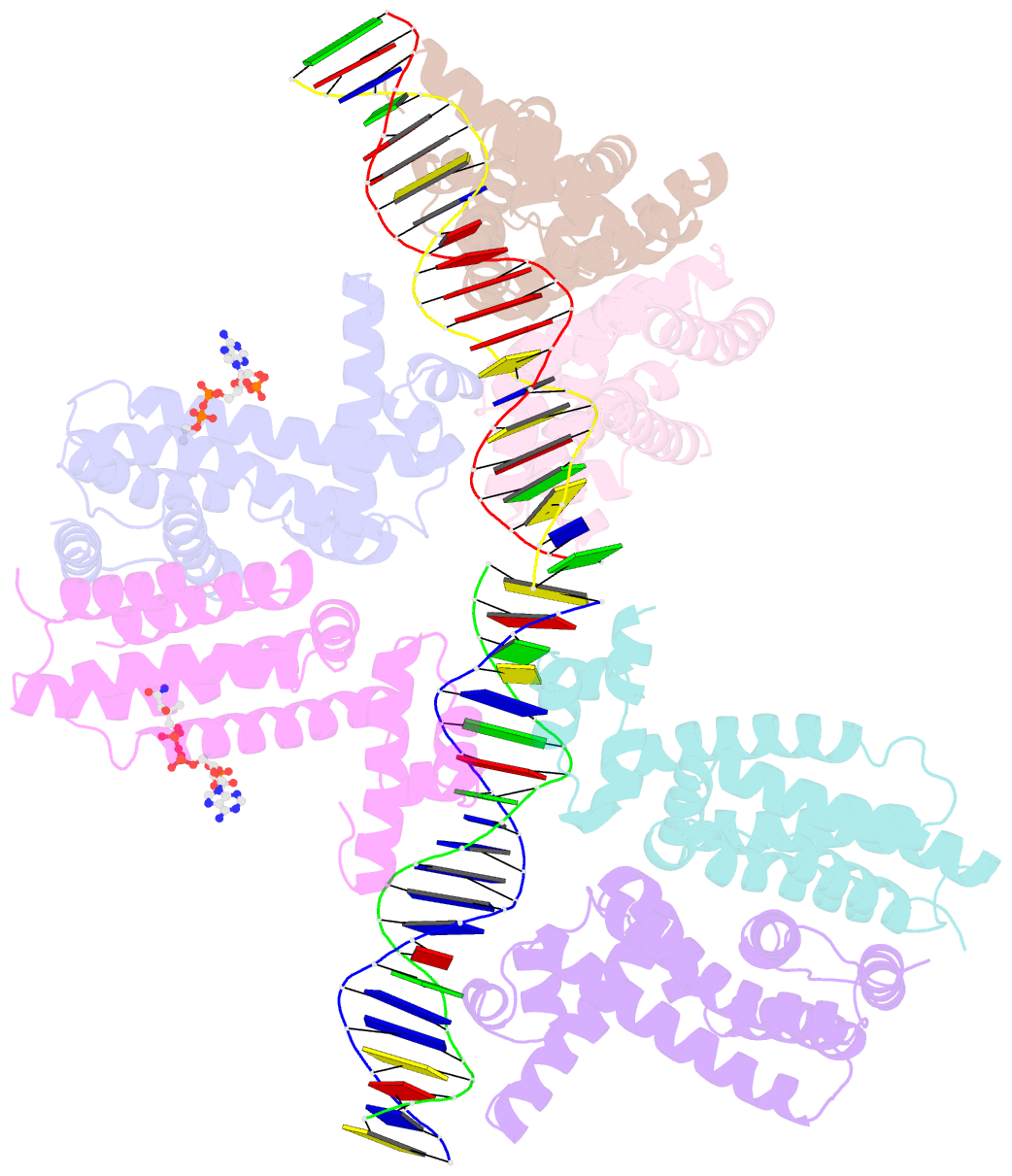
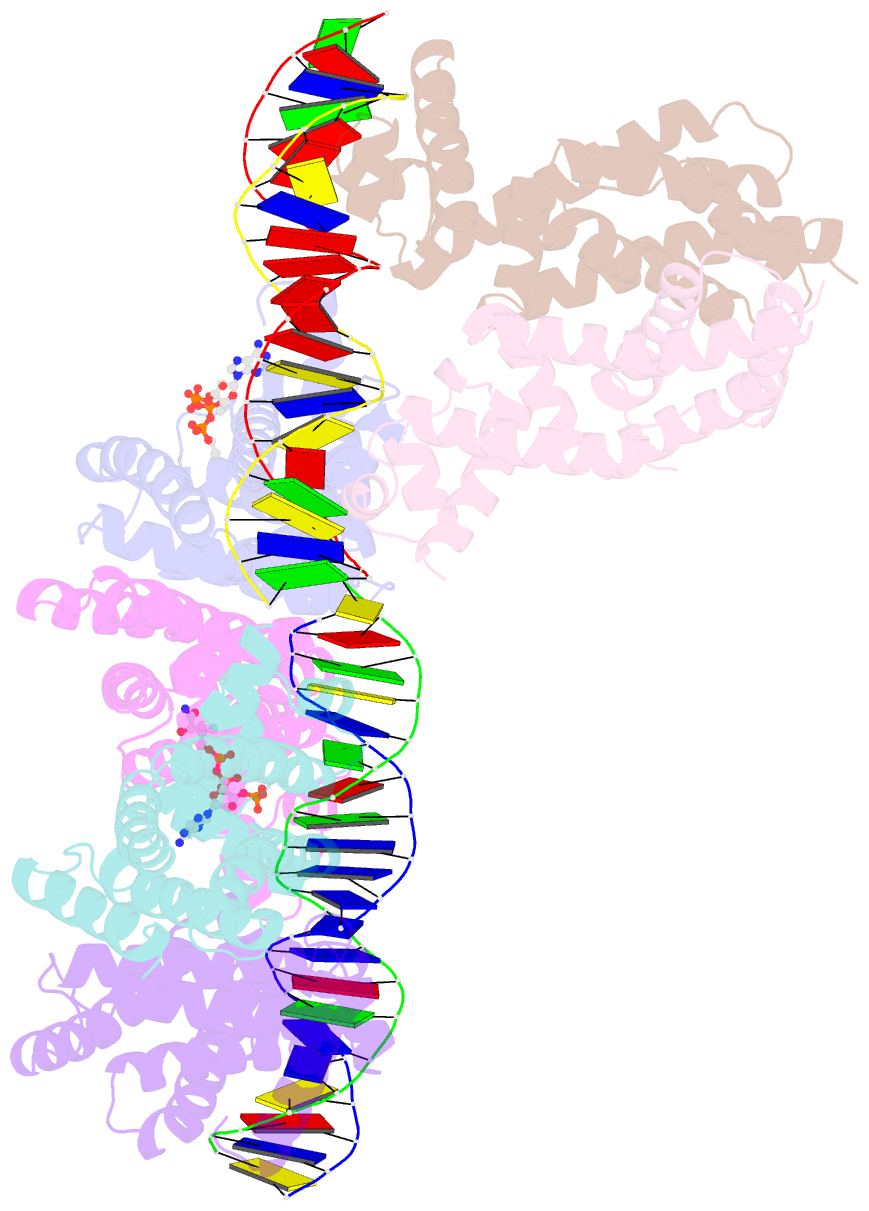
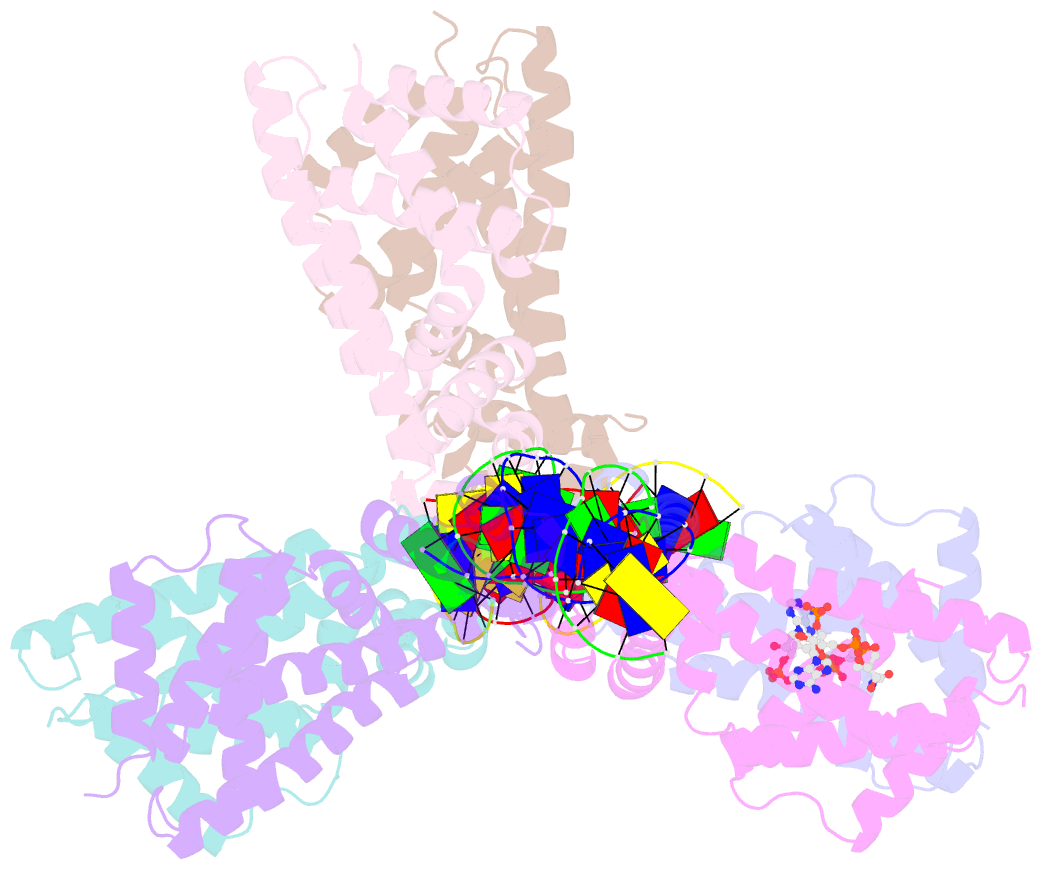
- Safadr in complex with dsDNA
- Reference
- Wang K, Sybers D, Maklad HR, Lemmens L, Lewyllie C, Zhou X, Schult F, Brasen C, Siebers B, Valegard K, Lindas AC, Peeters E (2019): "A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius." Nat Commun, 10, 1542. doi: 10.1038/s41467-019-09479-1.
- Abstract
- Fatty acid metabolism and its regulation are known to play important roles in bacteria and eukaryotes. By contrast, although certain archaea appear to metabolize fatty acids, the regulation of the underlying pathways in these organisms remains unclear. Here, we show that a TetR-family transcriptional regulator (FadRSa) is involved in regulation of fatty acid metabolism in the crenarchaeon Sulfolobus acidocaldarius. Functional and structural analyses show that FadRSa binds to DNA at semi-palindromic recognition sites in two distinct stoichiometric binding modes depending on the operator sequence. Genome-wide transcriptomic and chromatin immunoprecipitation analyses demonstrate that the protein binds to only four genomic sites, acting as a repressor of a 30-kb gene cluster comprising 23 open reading frames encoding lipases and β-oxidation enzymes. Fatty acyl-CoA molecules cause dissociation of FadRSa binding by inducing conformational changes in the protein. Our results indicate that, despite its similarity in overall structure to bacterial TetR-family FadR regulators, FadRSa displays a different acyl-CoA binding mode and a distinct regulatory mechanism.