Summary information and primary citation
- PDB-id
- 6exn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- cryo-EM (3.7 Å)
- Summary
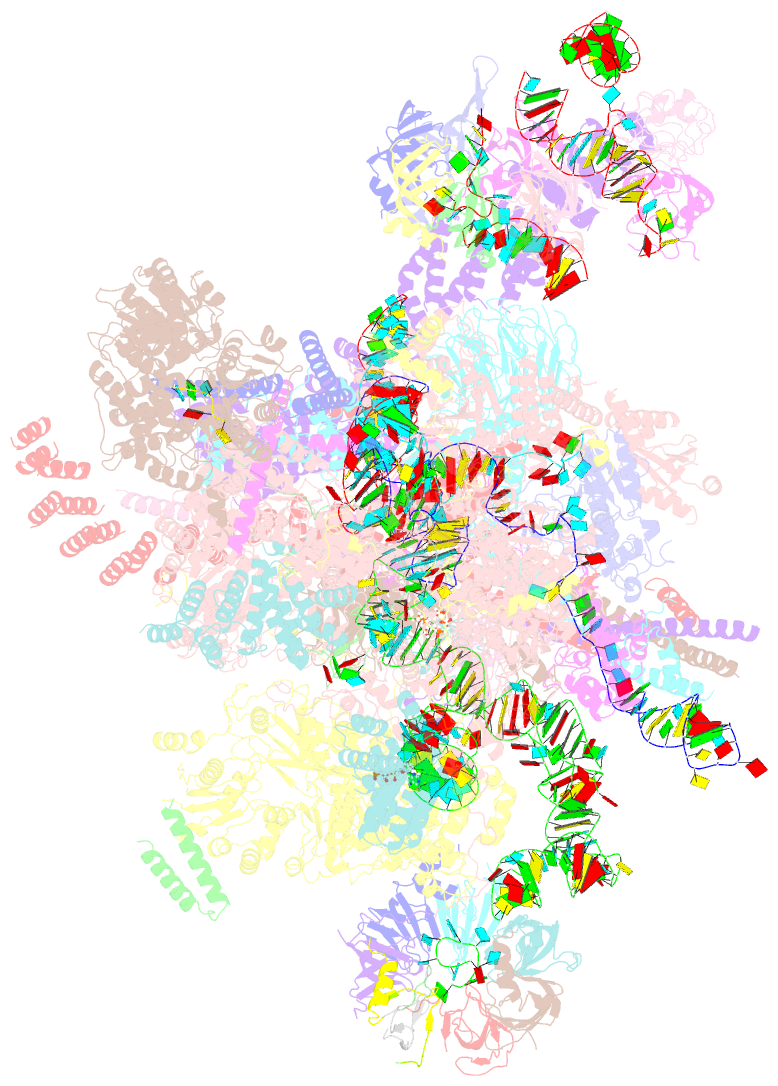
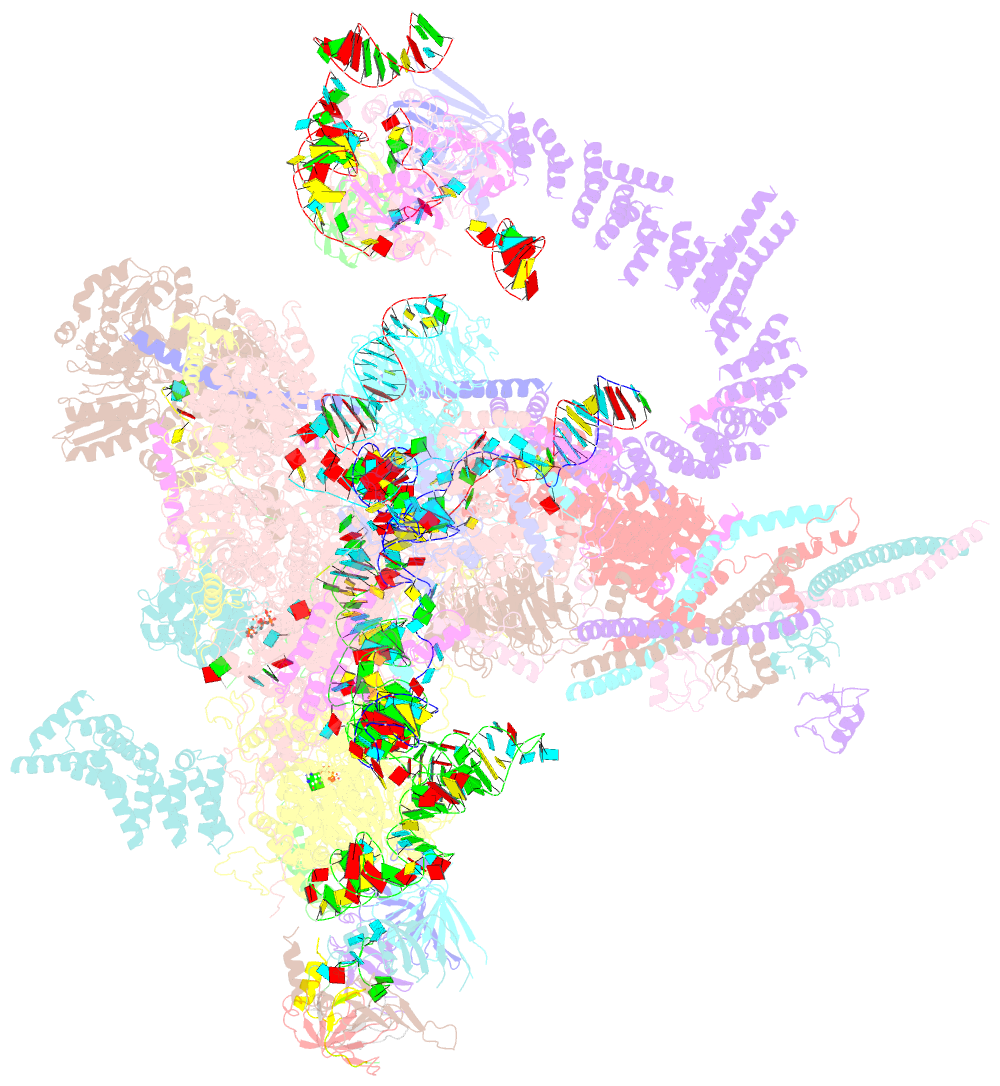
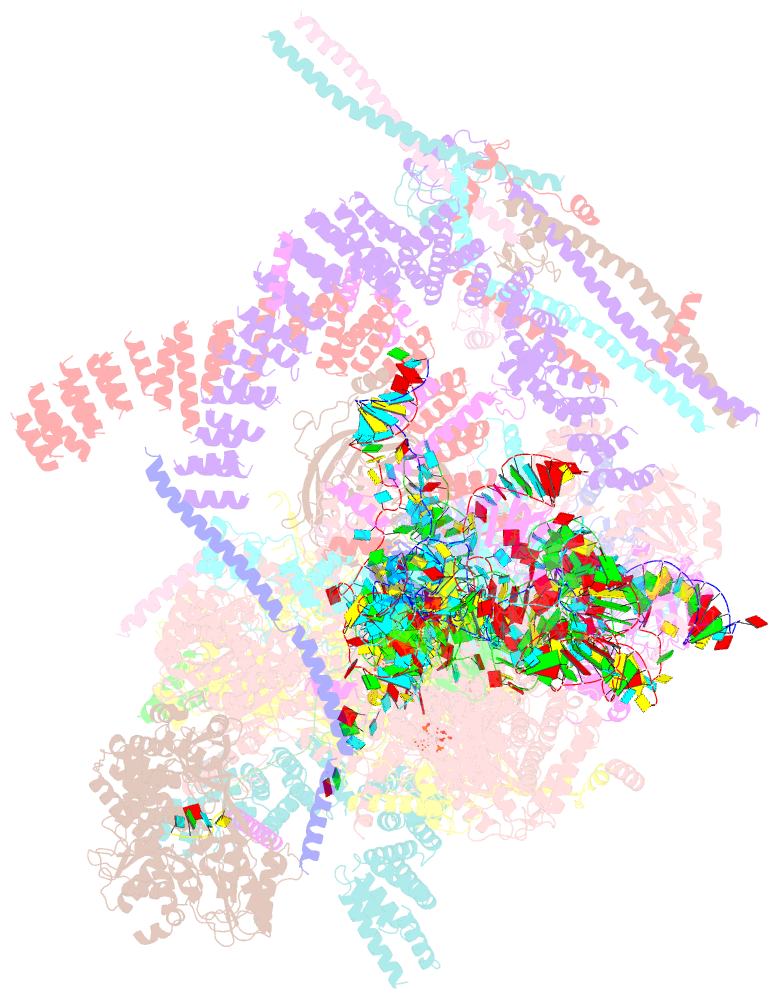
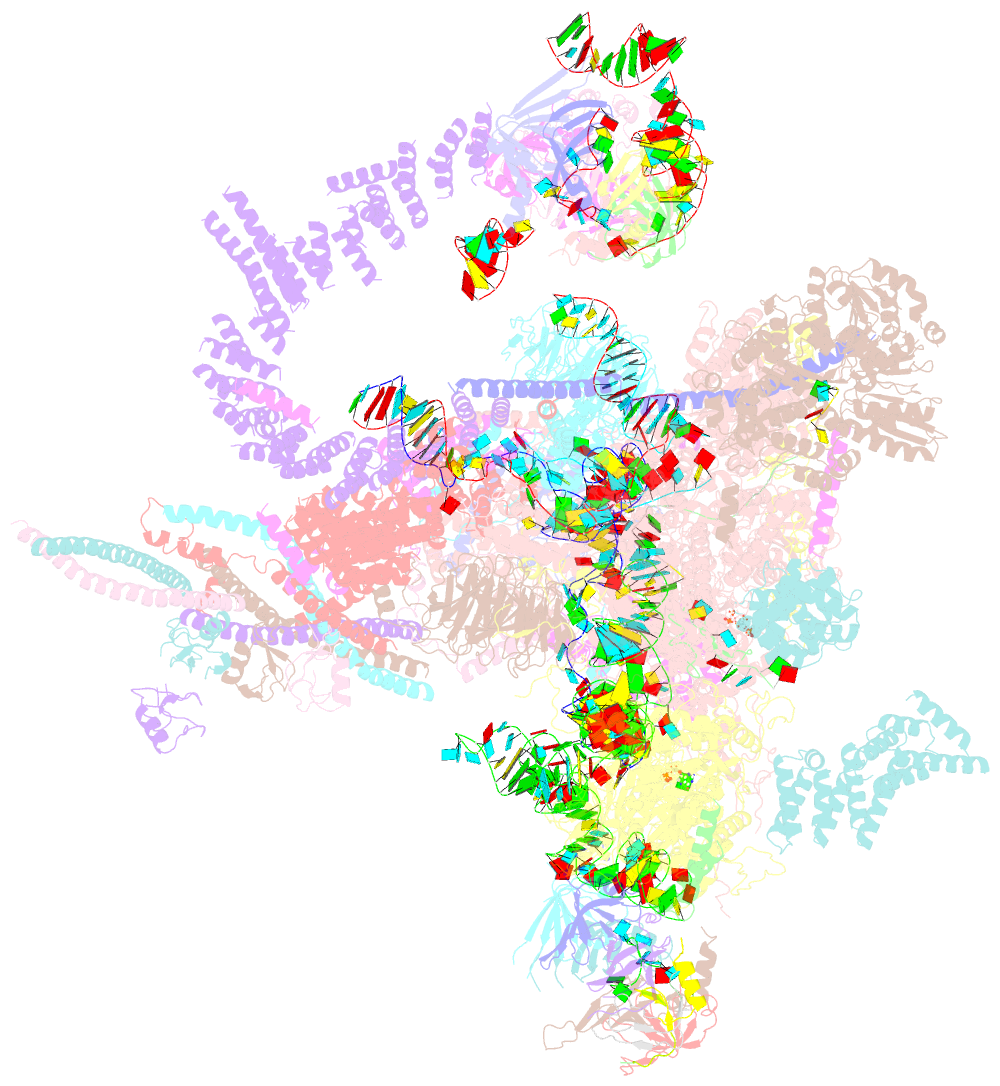
- Post-catalytic p complex spliceosome with 3' splice site docked
- Reference
- Wilkinson ME, Fica SM, Galej WP, Norman CM, Newman AJ, Nagai K (2017): "Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection." Science, 358, 1283-1288. doi: 10.1126/science.aar3729.
- Abstract
- Introns are removed from eukaryotic messenger RNA precursors by the spliceosome in two transesterification reactions-branching and exon ligation. The mechanism of 3'-splice site recognition during exon ligation has remained unclear. Here we present the 3.7-angstrom cryo-electron microscopy structure of the yeast P-complex spliceosome immediately after exon ligation. The 3'-splice site AG dinucleotide is recognized through non-Watson-Crick pairing with the 5' splice site and the branch-point adenosine. After the branching reaction, protein factors work together to remodel the spliceosome and stabilize a conformation competent for 3'-splice site docking, thereby promoting exon ligation. The structure accounts for the strict conservation of the GU and AG dinucleotides at the 5' and 3' ends of introns and provides insight into the catalytic mechanism of exon ligation.