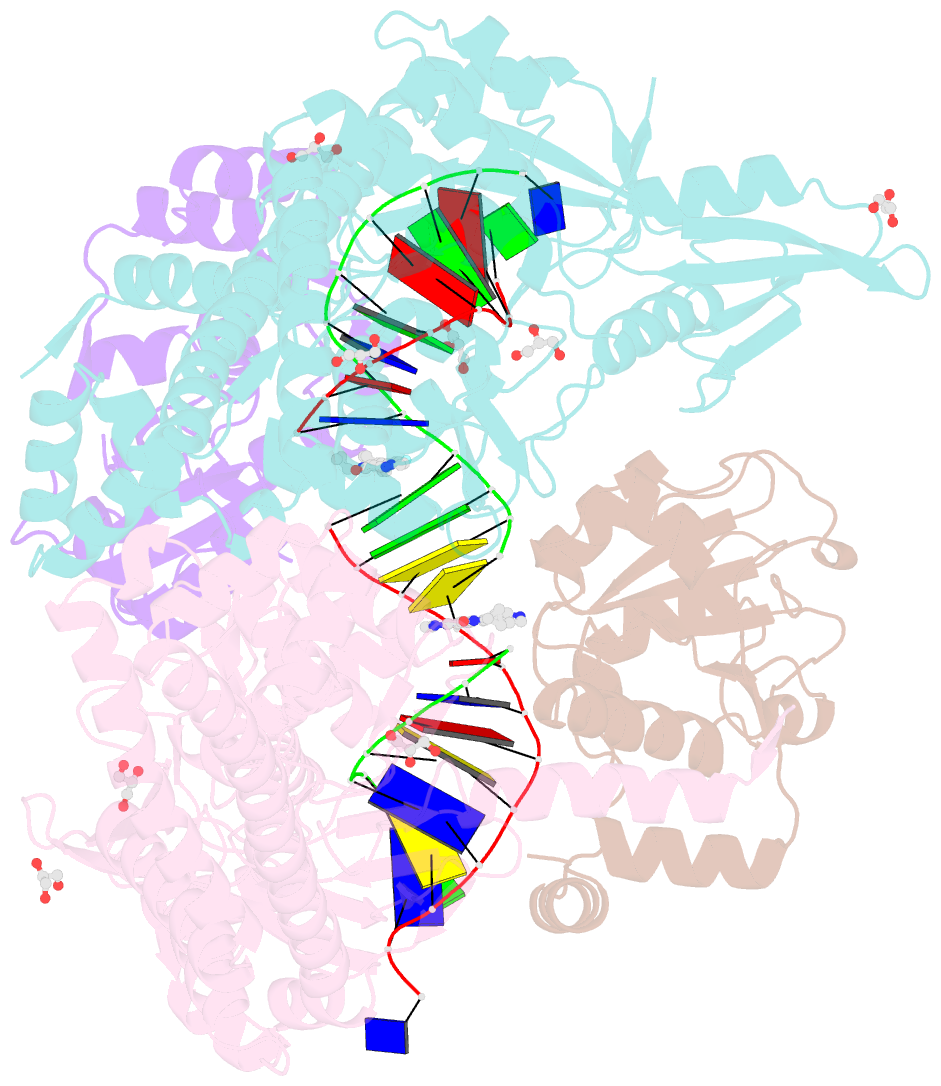
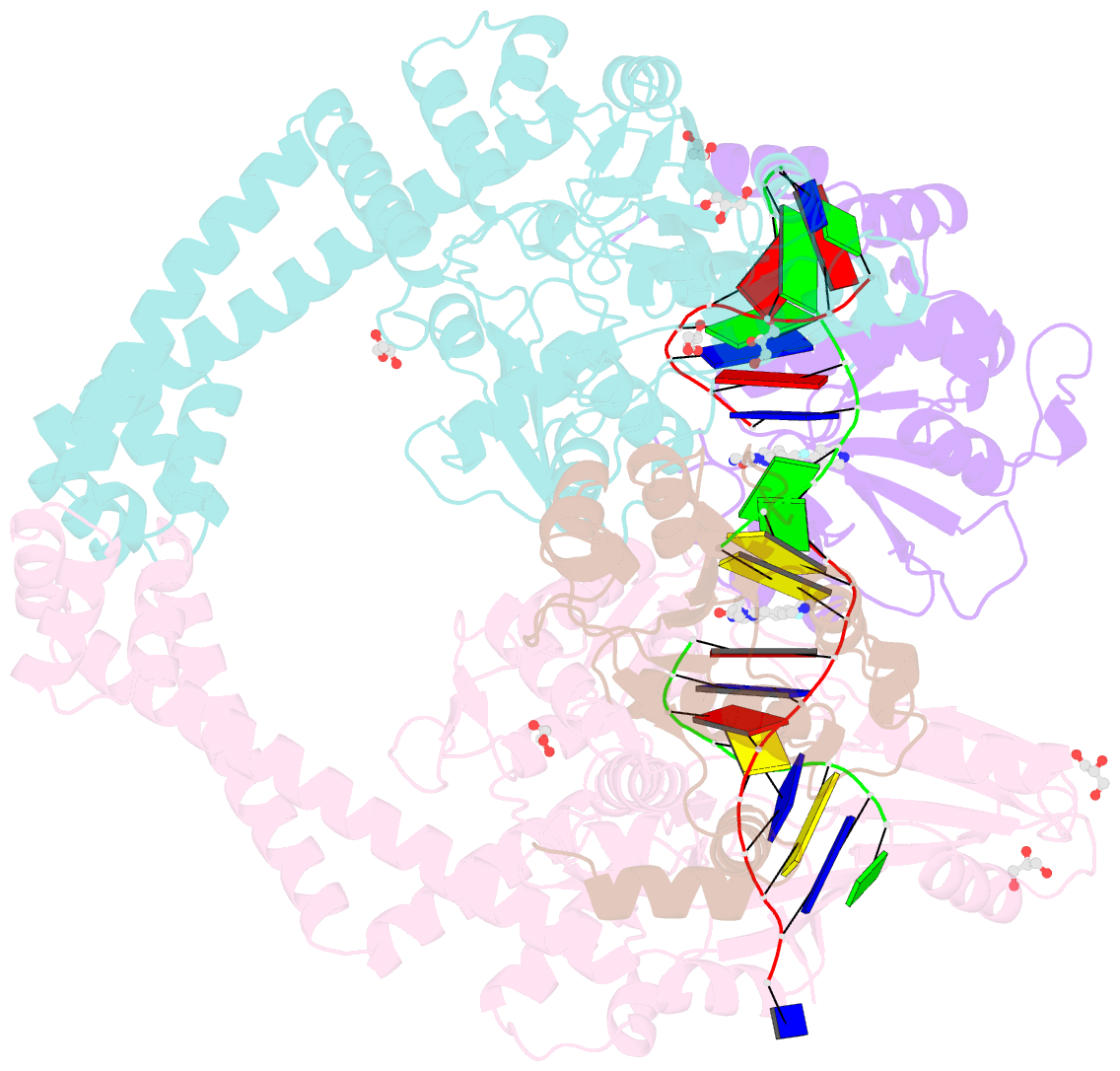
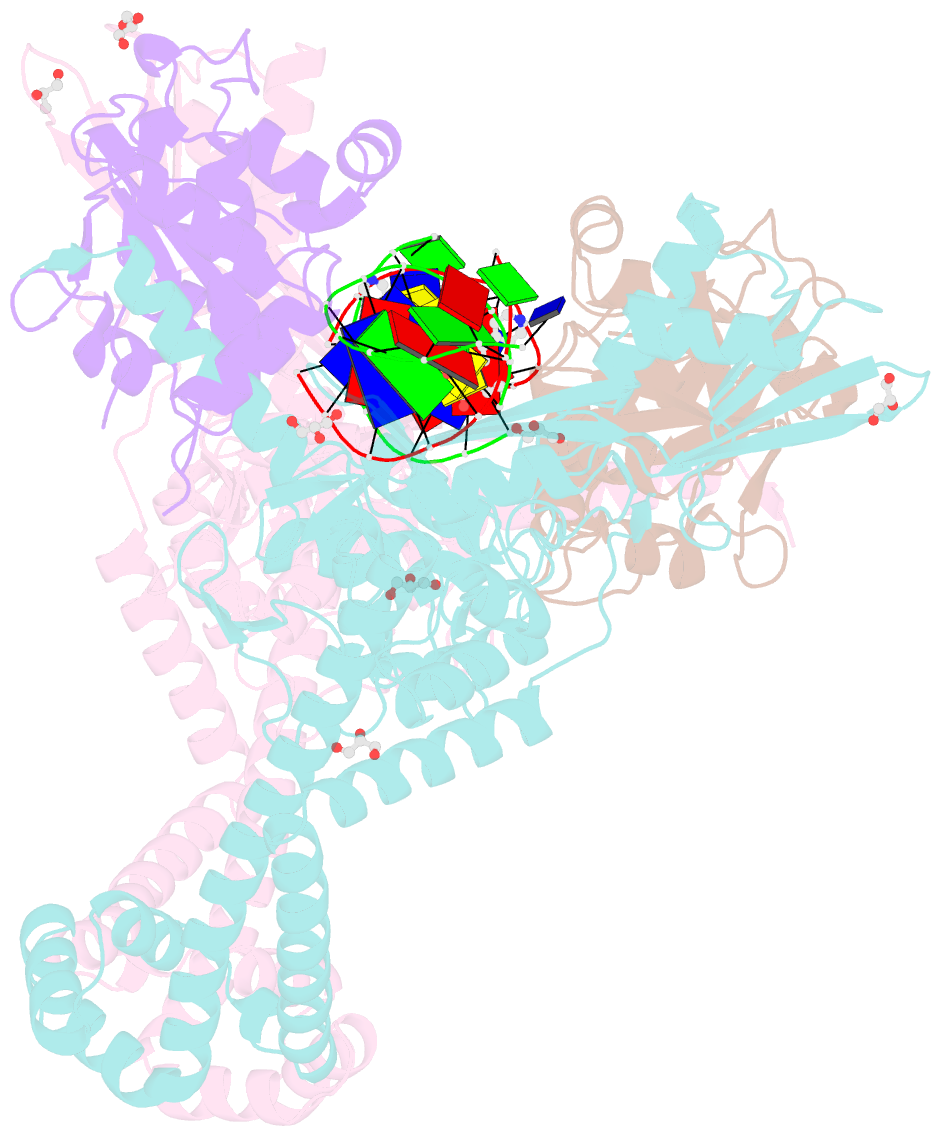
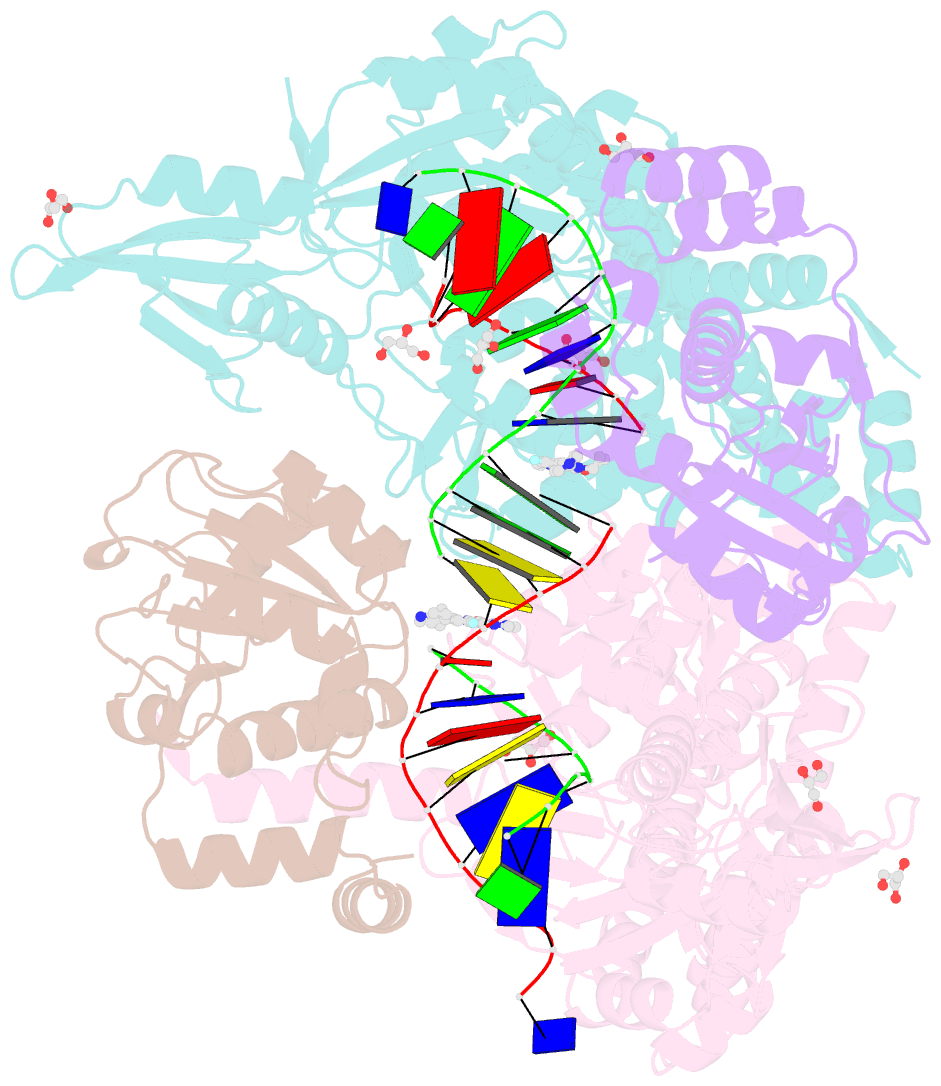
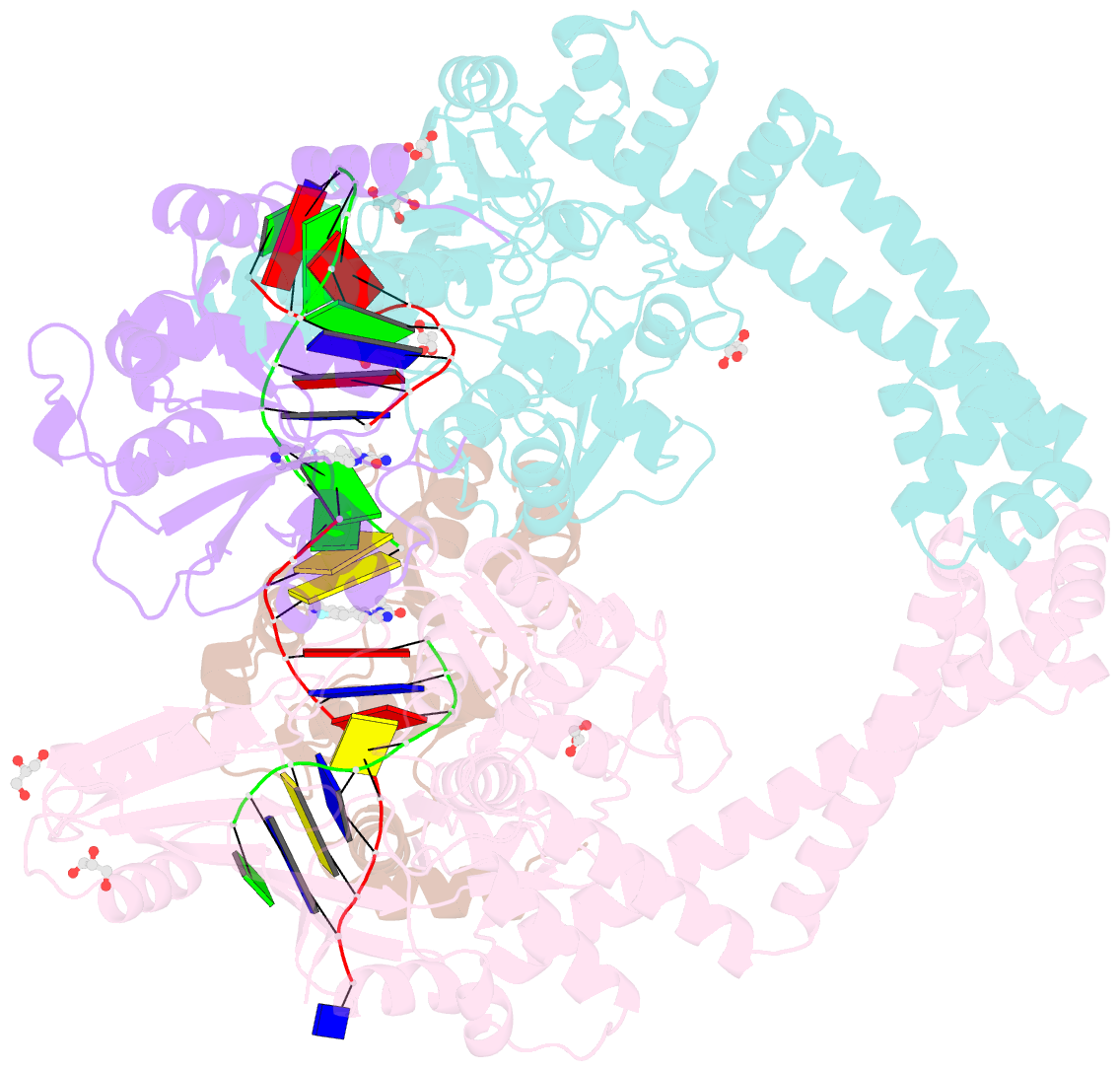
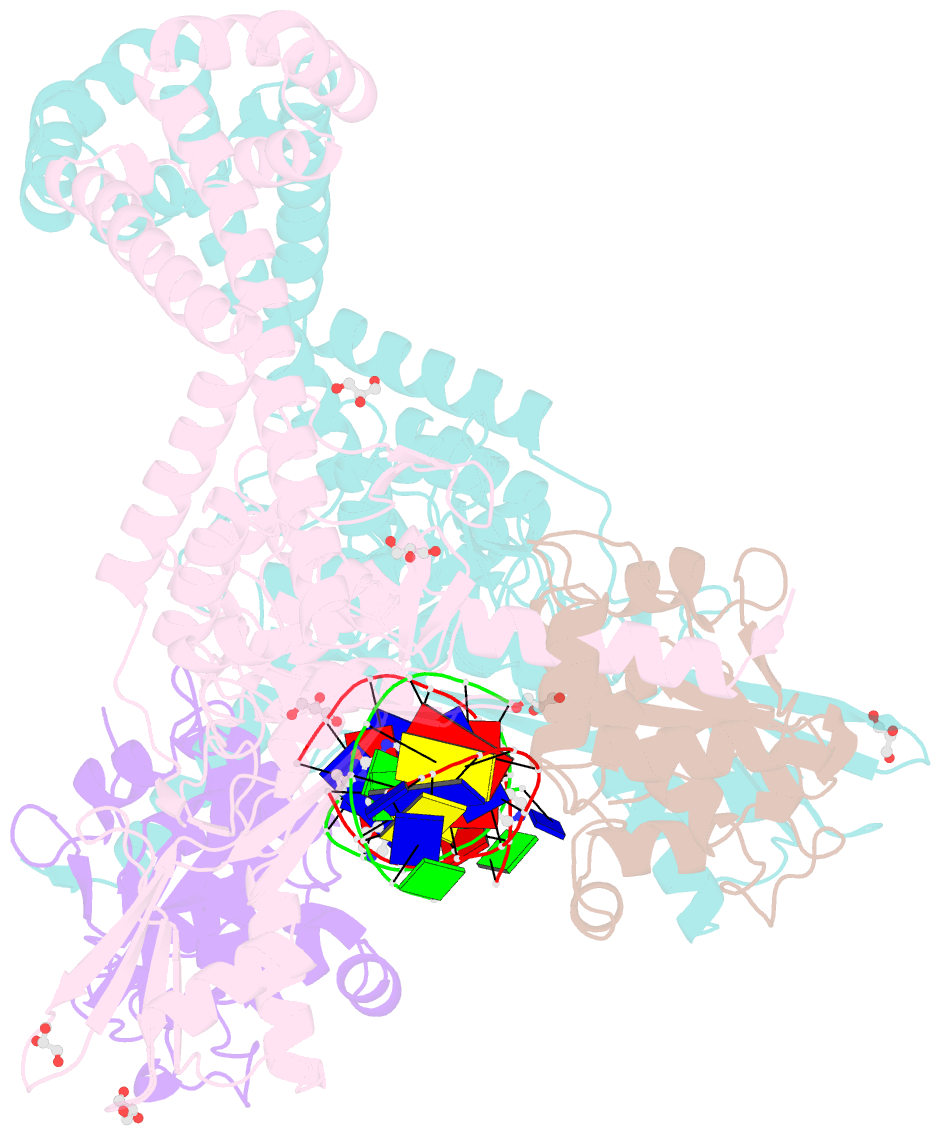
Summary information and primary citation
- PDB-id
- 6fqs; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase
- Method
- X-ray (3.11 Å)
- Summary
- 3.11a complex of s.aureus gyrase with imidazopyrazinone t3 and DNA
- Reference
- Germe T, Voros J, Jeannot F, Taillier T, Stavenger RA, Bacque E, Maxwell A, Bax BD (2018): "A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance." Nucleic Acids Res., 46, 4114-4128. doi: 10.1093/nar/gky181.
- Abstract
- Imidazopyrazinones (IPYs) are a new class of compounds that target bacterial topoisomerases as a basis for their antibacterial activity. We have characterized the mechanism of these compounds through structural/mechanistic studies showing they bind and stabilize a cleavage complex between DNA gyrase and DNA ('poisoning') in an analogous fashion to fluoroquinolones, but without the requirement for the water-metal-ion bridge. Biochemical experiments and structural studies of cleavage complexes of IPYs compared with an uncleaved gyrase-DNA complex, reveal conformational transitions coupled to DNA cleavage at the DNA gate. These involve movement at the GyrA interface and tilting of the TOPRIM domains toward the scissile phosphate coupled to capture of the catalytic metal ion. Our experiments show that these structural transitions are involved generally in poisoning of gyrase by therapeutic compounds and resemble those undergone by the enzyme during its adenosine triphosphate-coupled strand-passage cycle. In addition to resistance mutations affecting residues that directly interact with the compounds, we characterized a mutant (D82N) that inhibits formation of the cleavage complex by the unpoisoned enzyme. The D82N mutant appears to act by stabilizing the binary conformation of DNA gyrase with uncleaved DNA without direct interaction with the compounds. This provides general insight into the resistance mechanisms to antibiotics targeting bacterial type II topoisomerases.