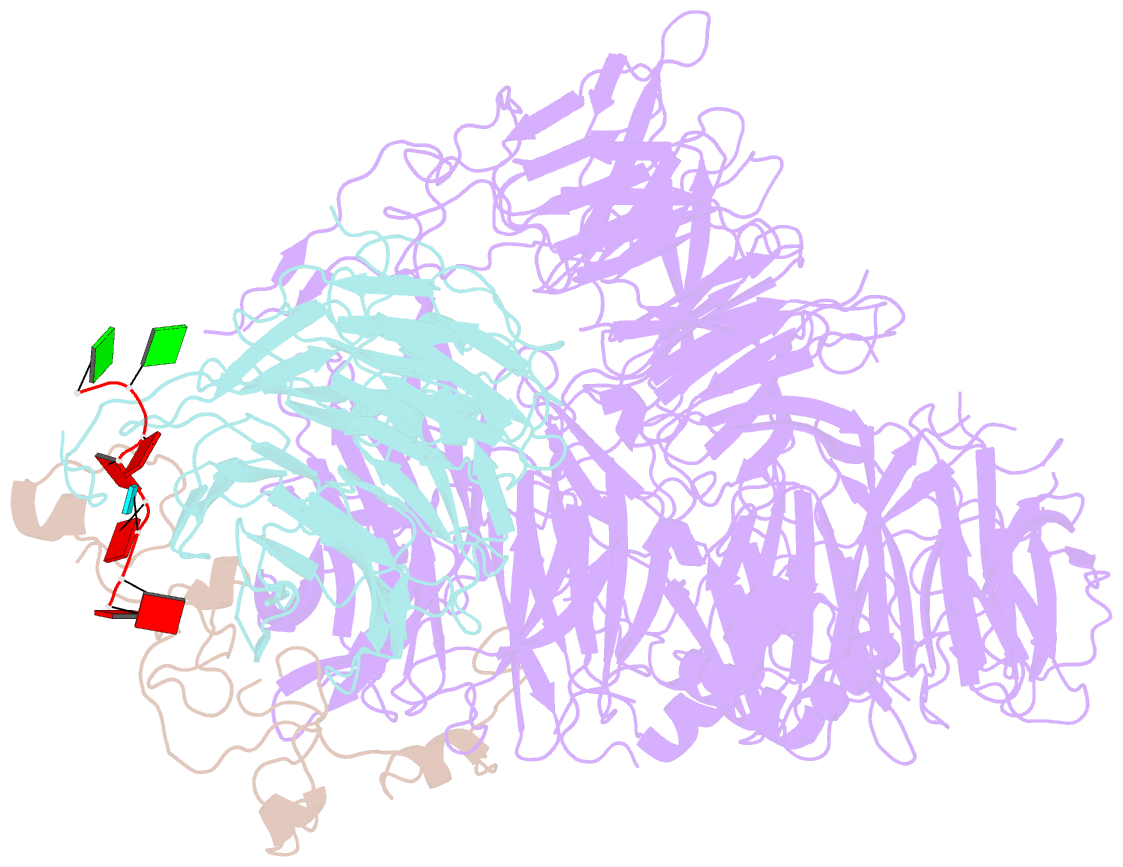
Summary information and primary citation
- PDB-id
- 6fuw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- cryo-EM (3.07 Å)
- Summary
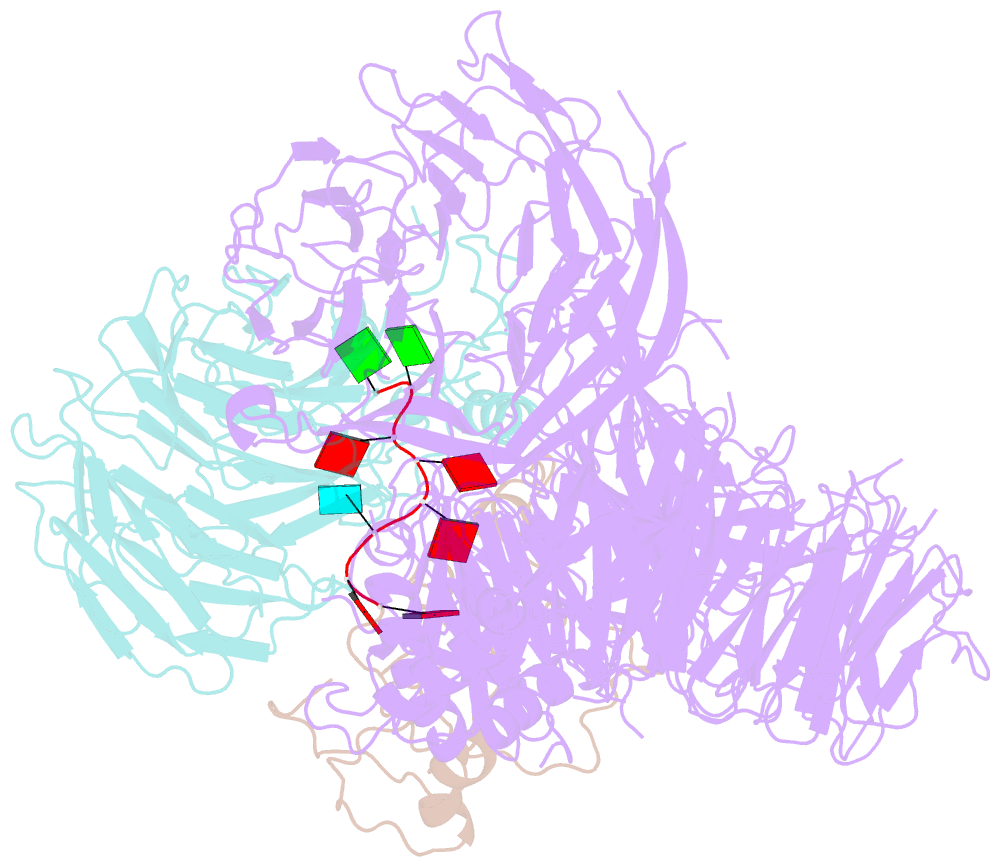
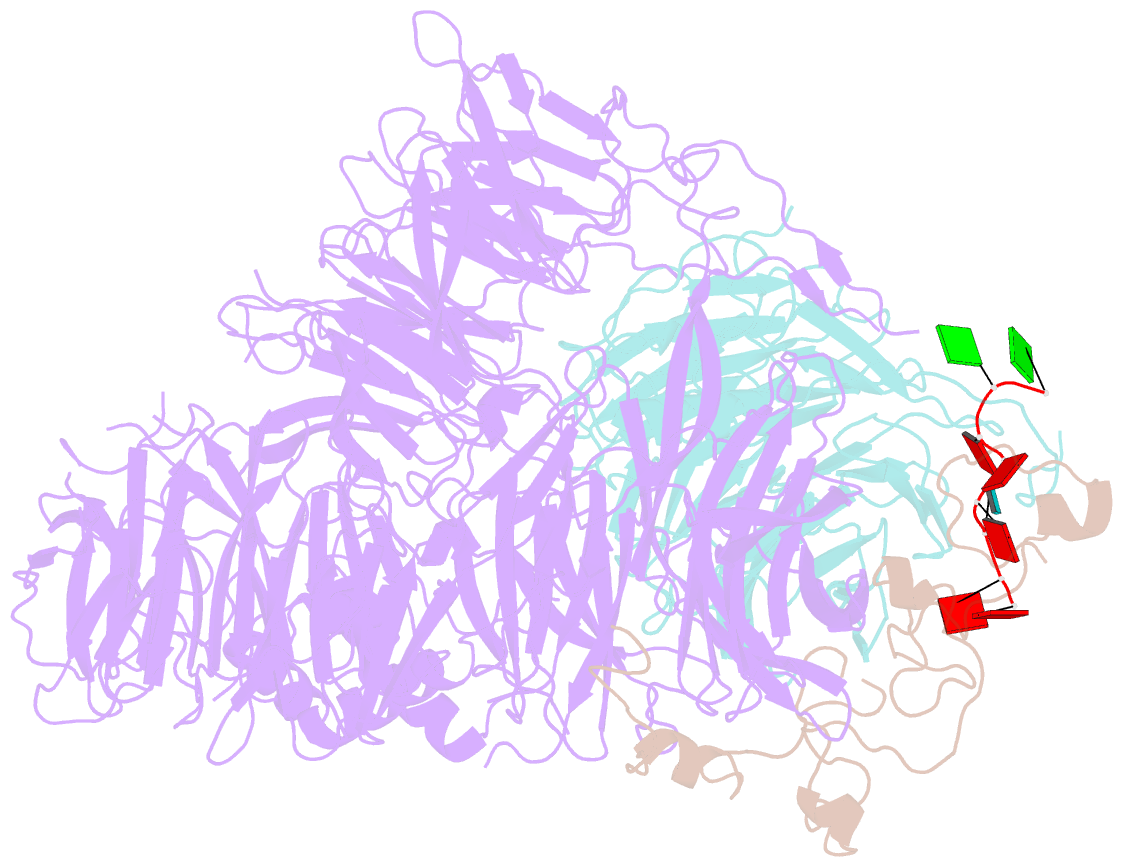
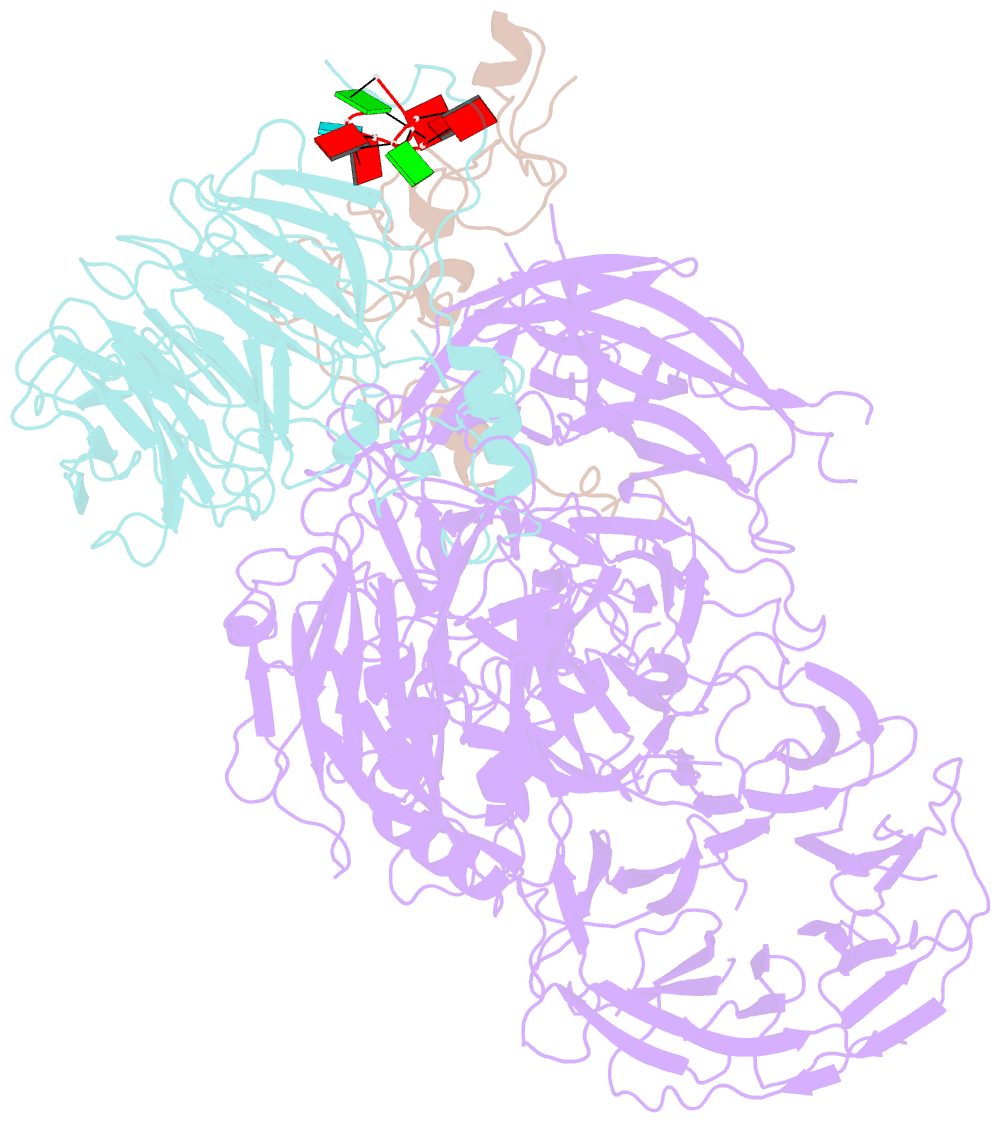
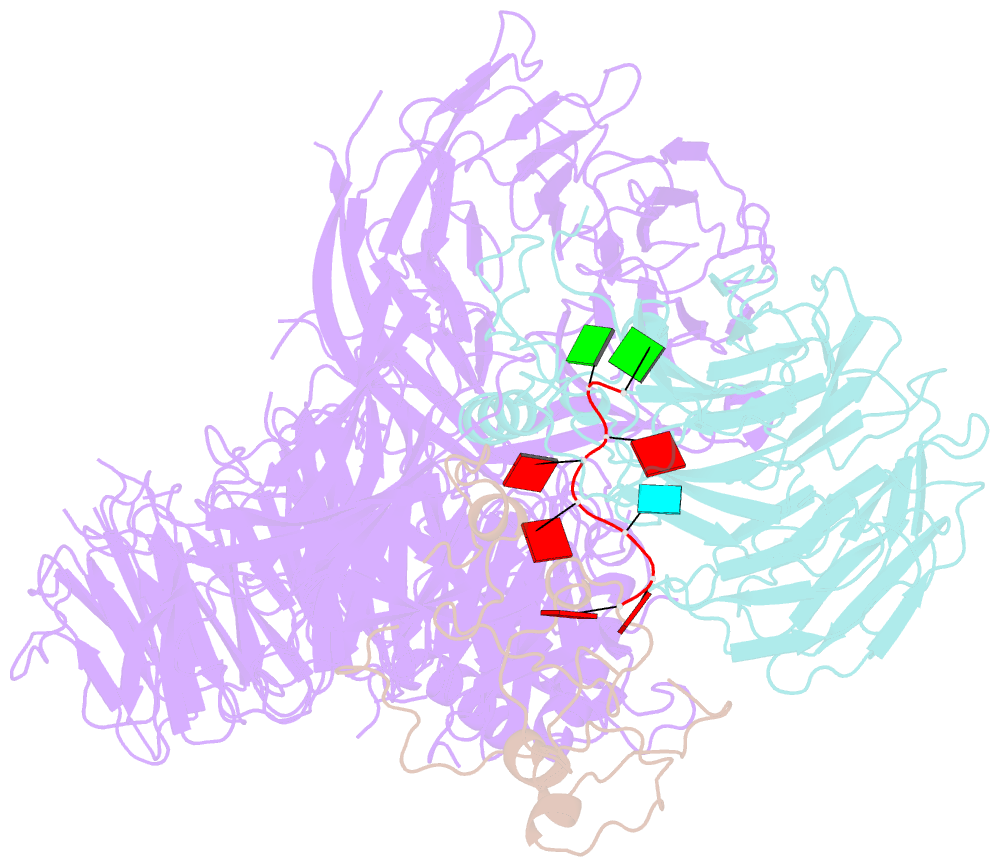
- cryo-EM structure of the human cpsf160-wdr33-cpsf30 complex bound to the pas aauaaa motif at 3.1 angstrom resolution
- Reference
- Clerici M, Faini M, Muckenfuss LM, Aebersold R, Jinek M (2018): "Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex." Nat. Struct. Mol. Biol., 25, 135-138. doi: 10.1038/s41594-017-0020-6.
- Abstract
- Mammalian mRNA biogenesis requires specific recognition of a hexanucleotide AAUAAA motif in the polyadenylation signals (PAS) of precursor mRNA (pre-mRNA) transcripts by the cleavage and polyadenylation specificity factor (CPSF) complex. Here we present a 3.1-Å-resolution cryo-EM structure of a core CPSF module bound to the PAS hexamer motif. The structure reveals the molecular interactions responsible for base-specific recognition, providing a rationale for mechanistic differences between mammalian and yeast 3' polyadenylation.