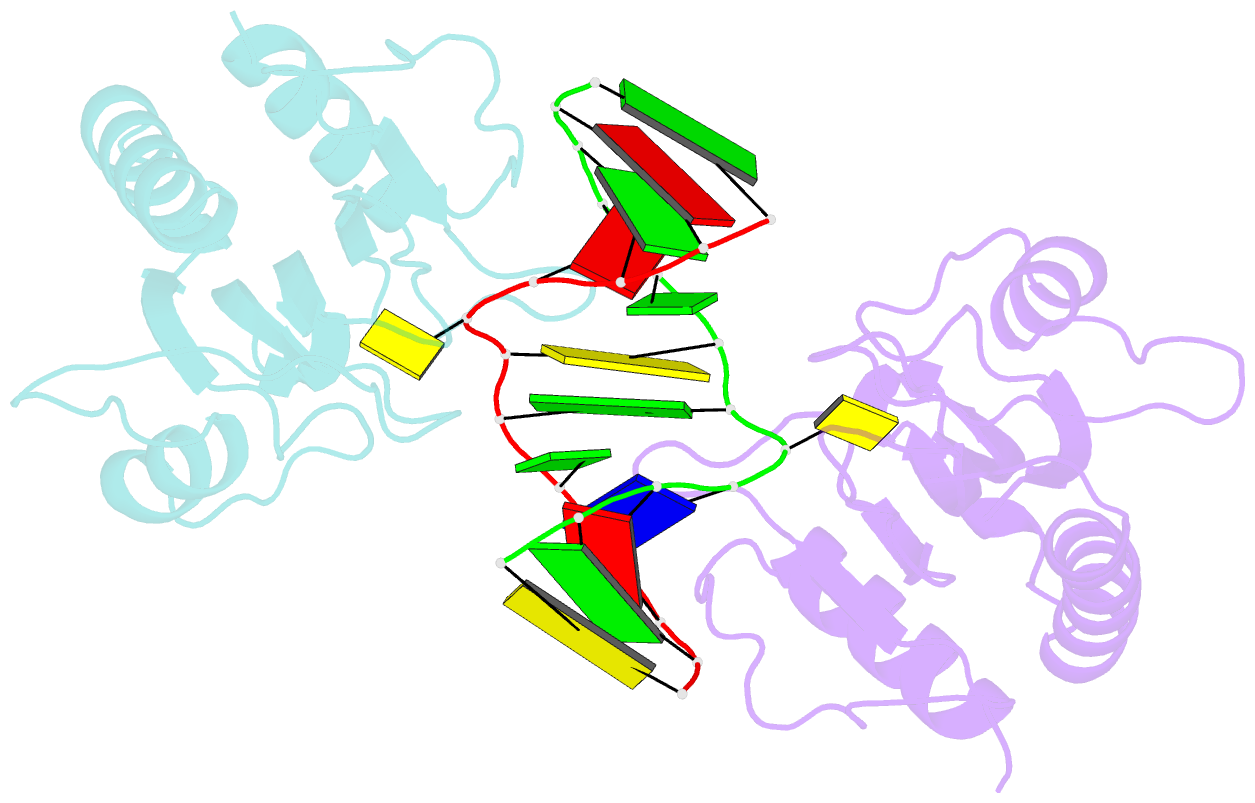
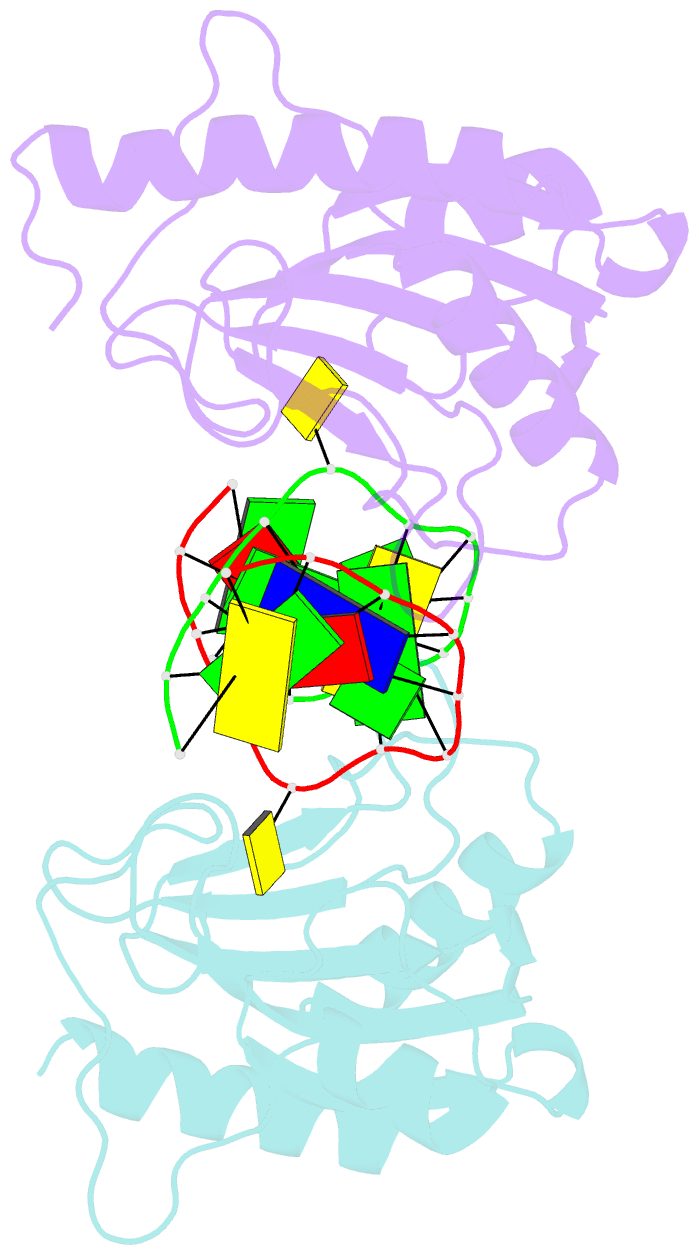
Summary information and primary citation
- PDB-id
- 6gcf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (1.55 Å)
- Summary
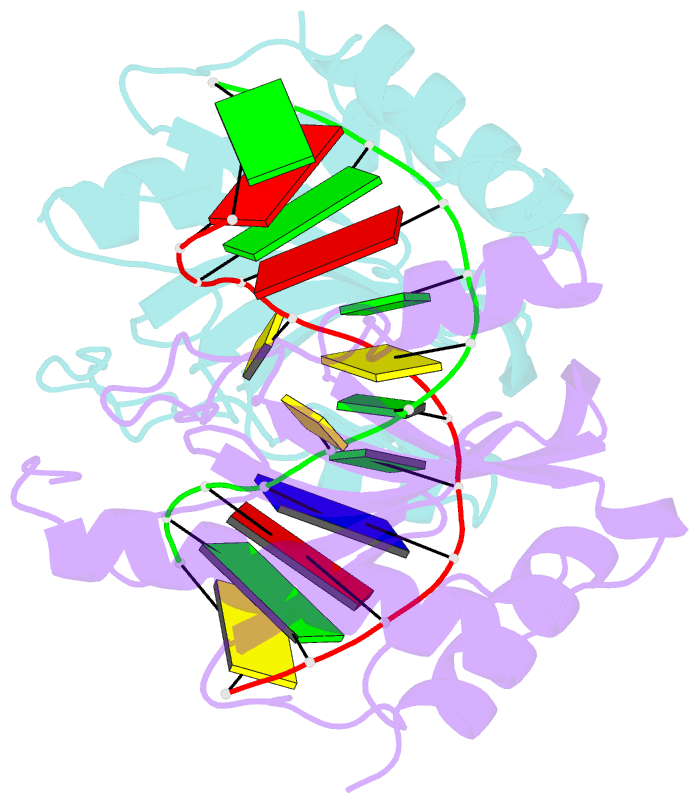
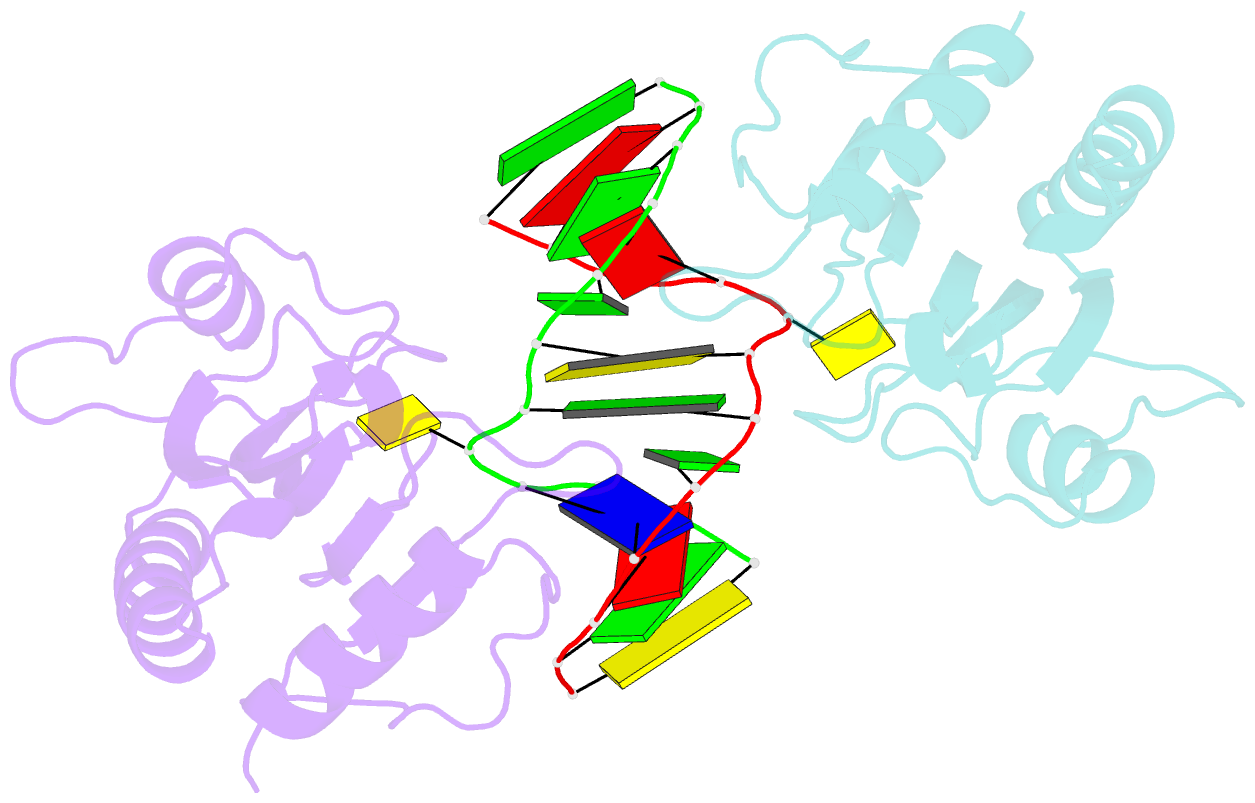
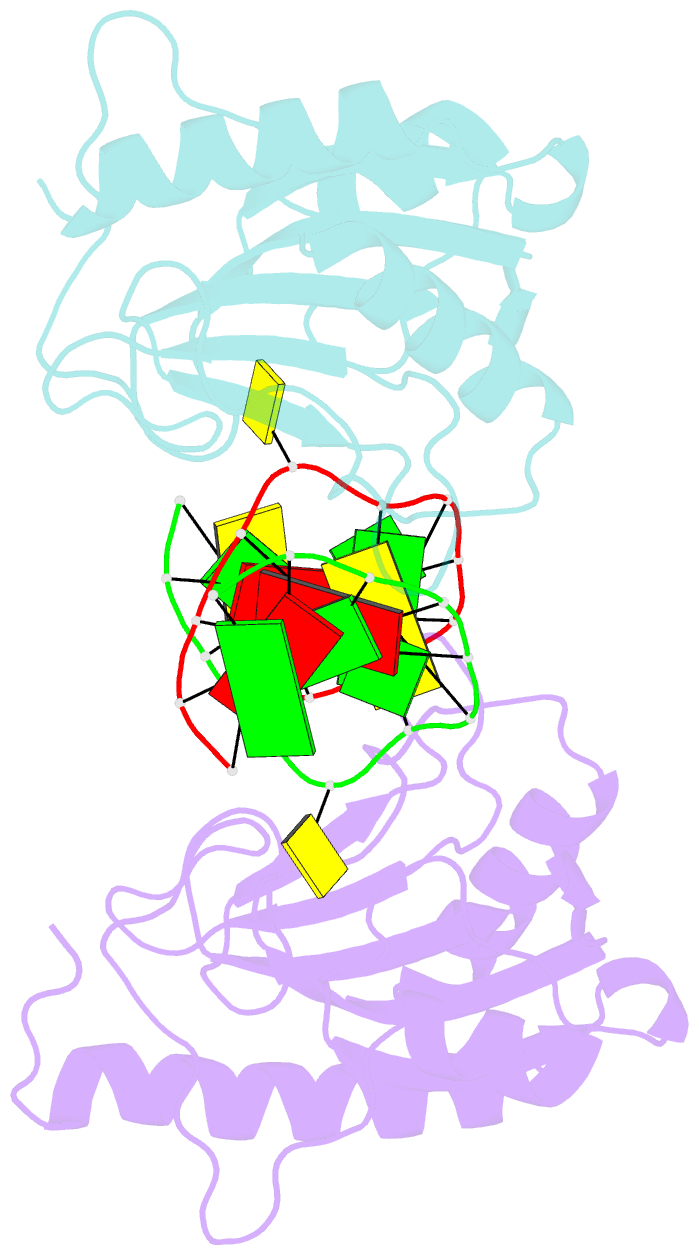
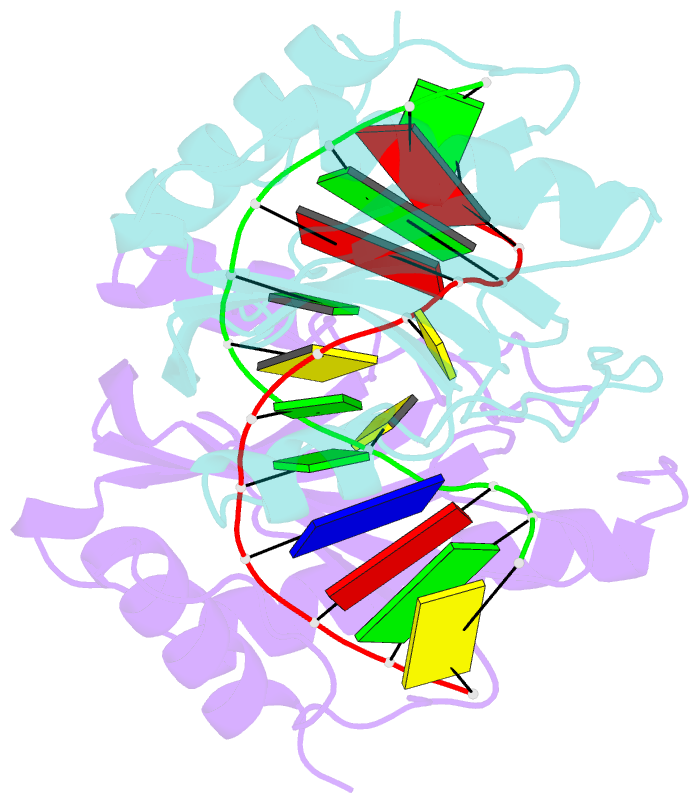
- DNA binding domain of restriction endonuclease mcrbc in complex with n4-methylcytosine DNA
- Reference
- Zagorskaite E, Manakova E, Sasnauskas G (2018): "Recognition of modified cytosine variants by the DNA-binding domain of methyl-directed endonuclease McrBC." FEBS Lett., 592, 3335-3345. doi: 10.1002/1873-3468.13244.
- Abstract
- Cytosine modifications expand the information content of genomic DNA in both eukaryotes and prokaryotes, providing means for epigenetic regulation and self versus nonself discrimination. For example, the methyl-directed restriction endonuclease, McrBC, recognizes and cuts invading bacteriophage DNA containing 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), and N4-methylcytosine (4mC), leaving the unmodified host DNA intact. Here, we present cocrystal structures of McrB-N bound to DNA oligoduplexes containing 5hmC, 5-formylcytosine (5fC), and 4mC, and characterize the relative affinity of McrB-N to various cytosine variants. We find that McrB-N flips out modified bases into a protein pocket and binds cytosine derivatives in the order of descending affinity: 4mC > 5mC > 5hmC ≫ 5fC. We also show that pocket mutations alter the relative preference of McrB-N to 5mC, 5hmC, and 4mC.