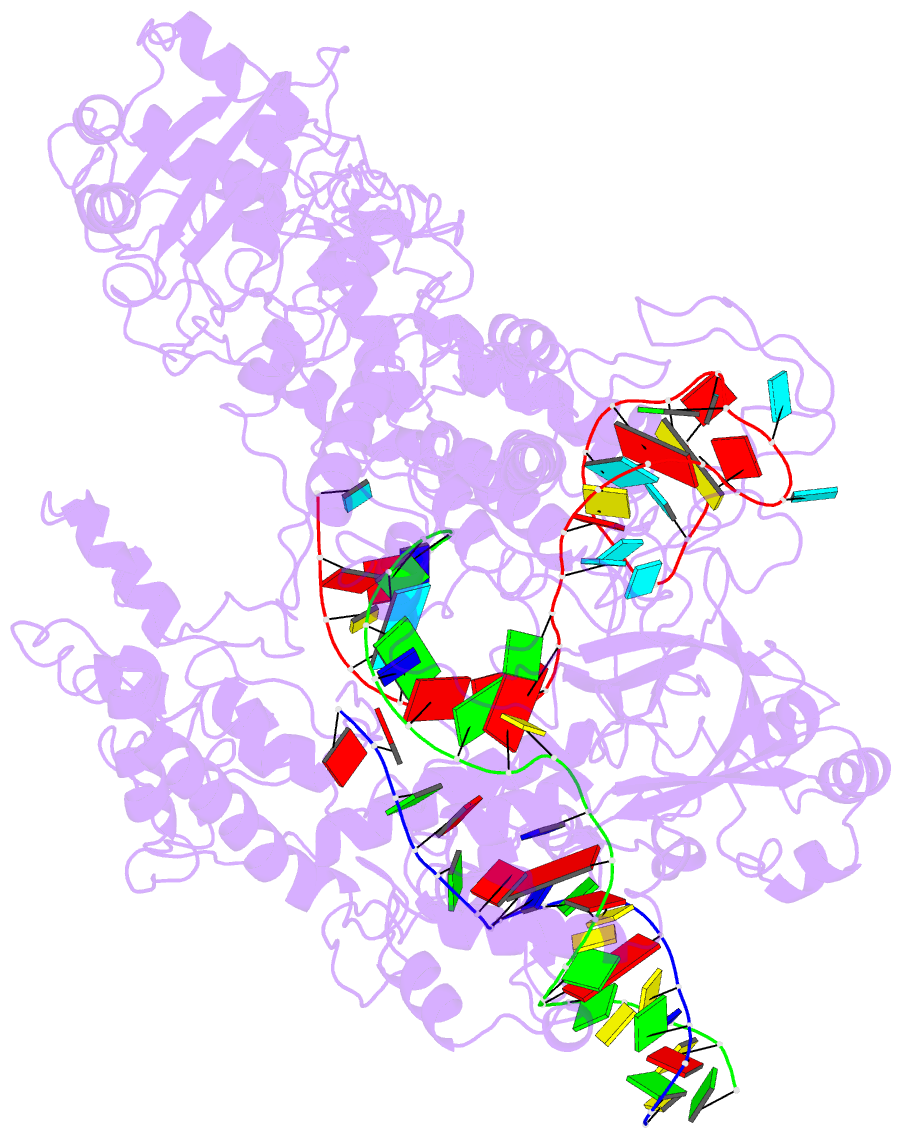
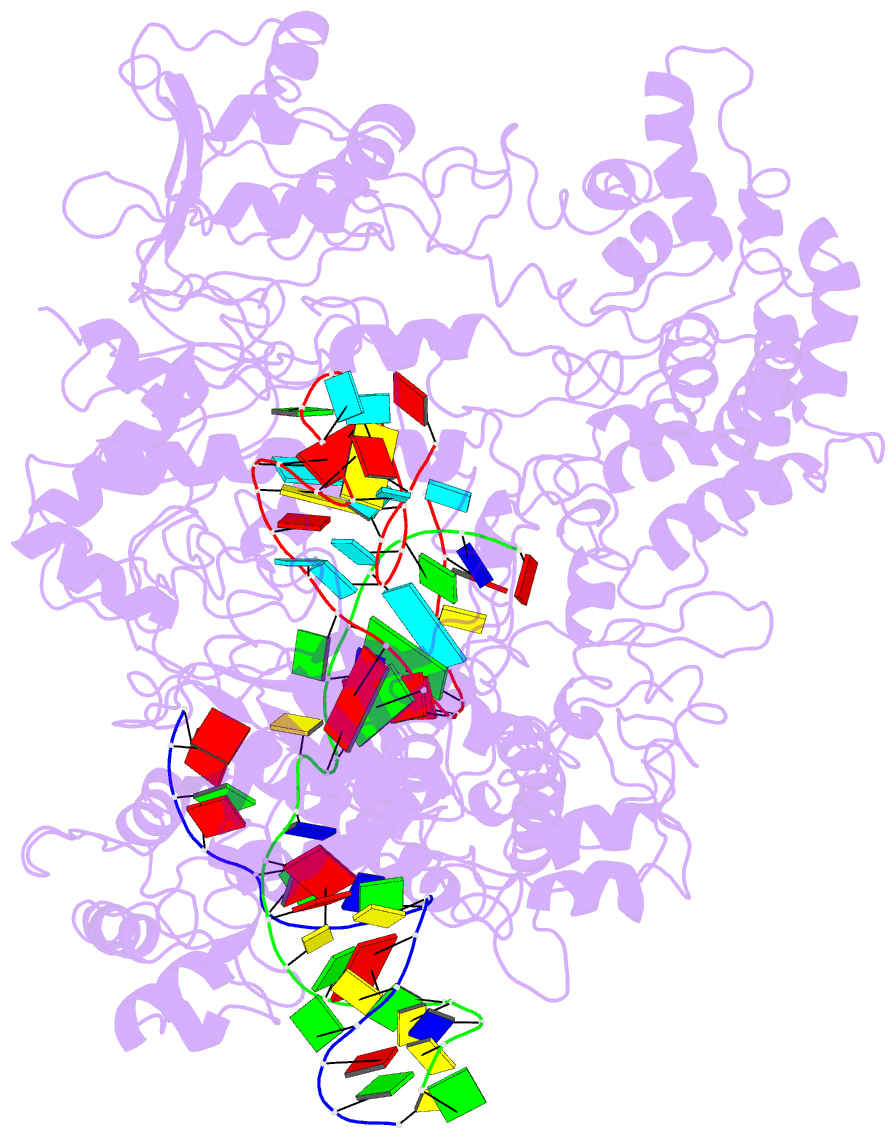
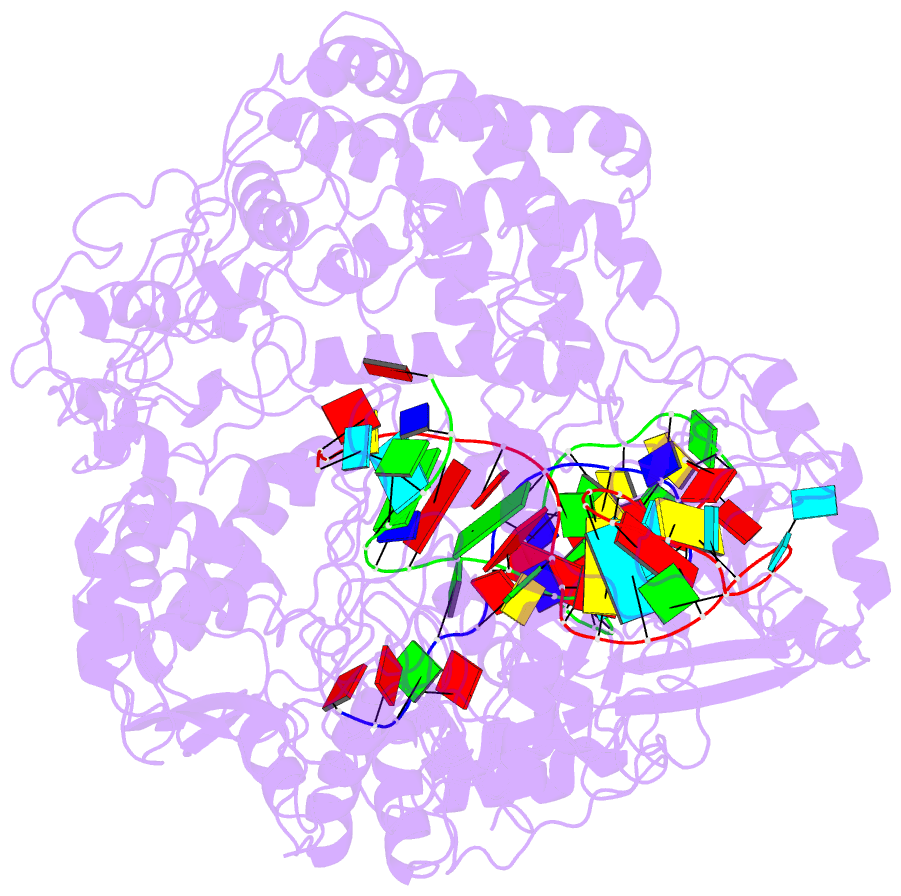
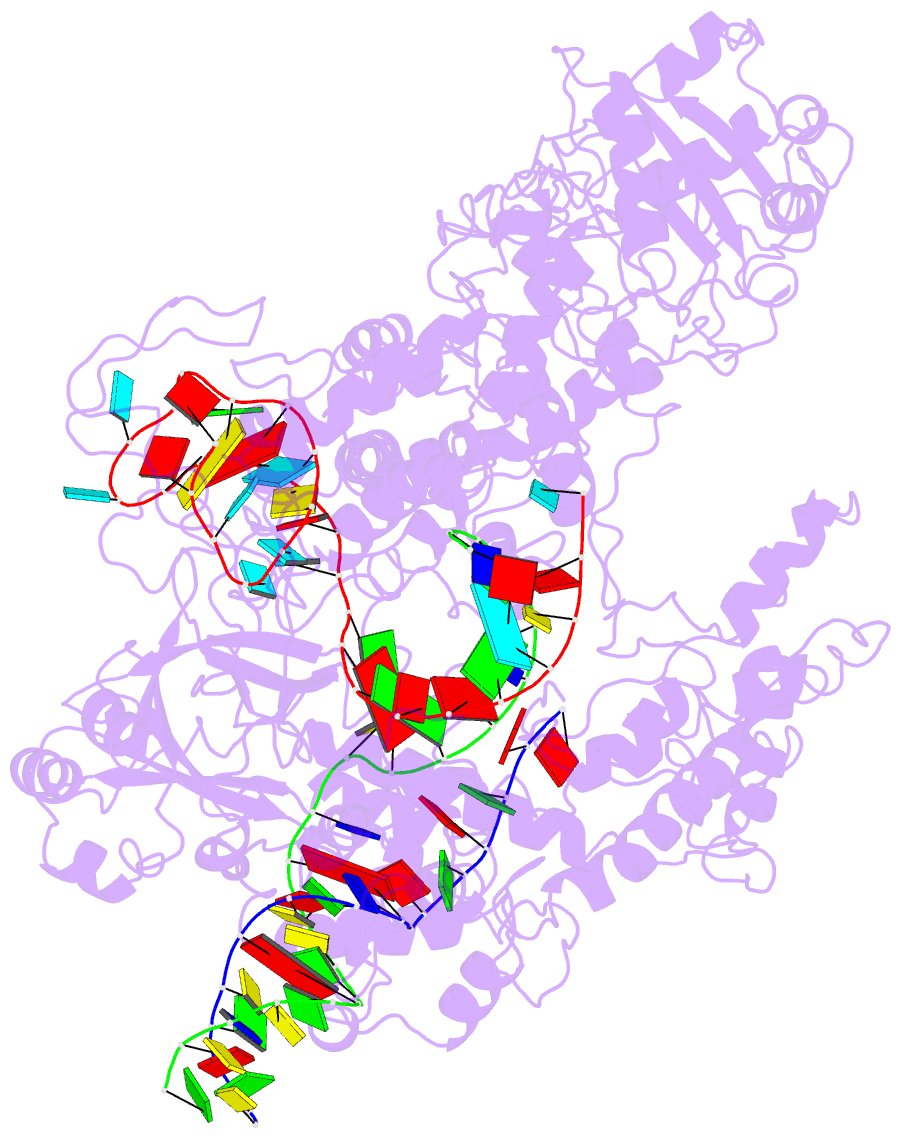
Summary information and primary citation
- PDB-id
- 6gtf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- cryo-EM (3.63 Å)
- Summary
- Transient state structure of crispr-cpf1 (cas12a) i5 conformation
- Reference
- Stella S, Mesa P, Thomsen J, Paul B, Alcon P, Jensen SB, Saligram B, Moses ME, Hatzakis NS, Montoya G (2018): "Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity." Cell, 175, 1856-1871.e21. doi: 10.1016/j.cell.2018.10.045.
- Abstract
- Cas12a, also known as Cpf1, is a type V-A CRISPR-Cas RNA-guided endonuclease that is used for genome editing based on its ability to generate specific dsDNA breaks. Here, we show cryo-EM structures of intermediates of the cleavage reaction, thus visualizing three protein regions that sense the crRNA-DNA hybrid assembly triggering the catalytic activation of Cas12a. Single-molecule FRET provides the thermodynamics and kinetics of the conformational activation leading to phosphodiester bond hydrolysis. These findings illustrate why Cas12a cuts its target DNA and unleashes unspecific cleavage activity, degrading ssDNA molecules after activation. In addition, we show that other crRNAs are able to displace the R-loop inside the protein after target DNA cleavage, terminating indiscriminate ssDNA degradation. We propose a model whereby the conformational activation of the enzyme results in indiscriminate ssDNA cleavage. The displacement of the R-loop by a new crRNA molecule will reset Cas12a specificity, targeting new DNAs.