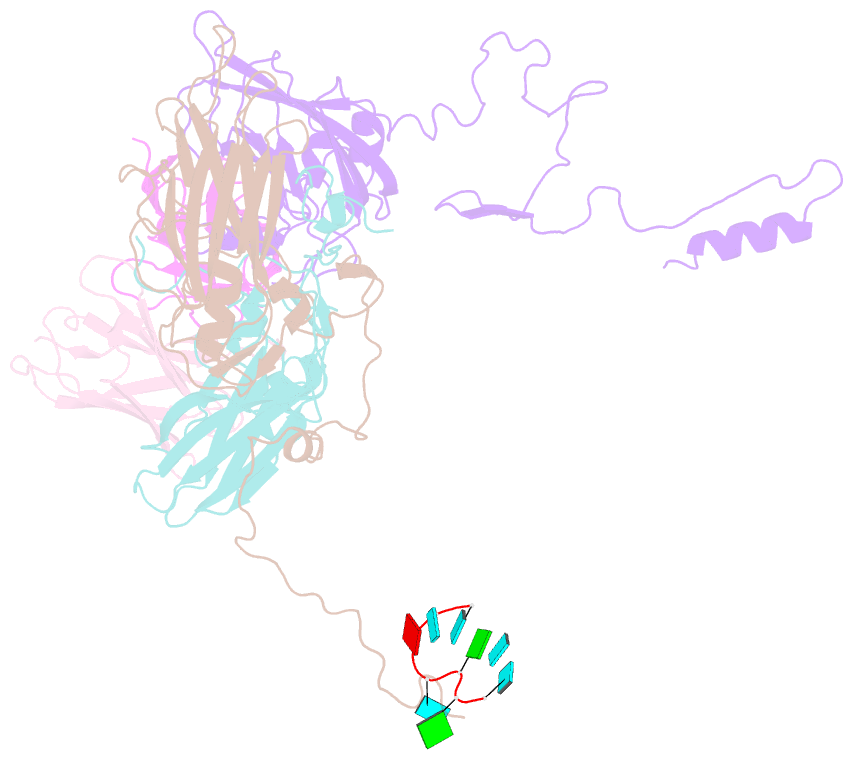
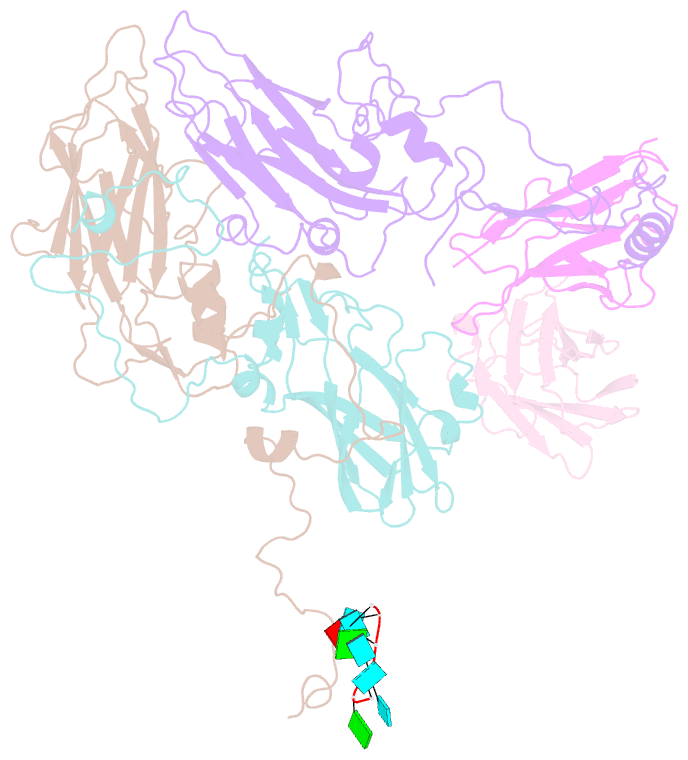
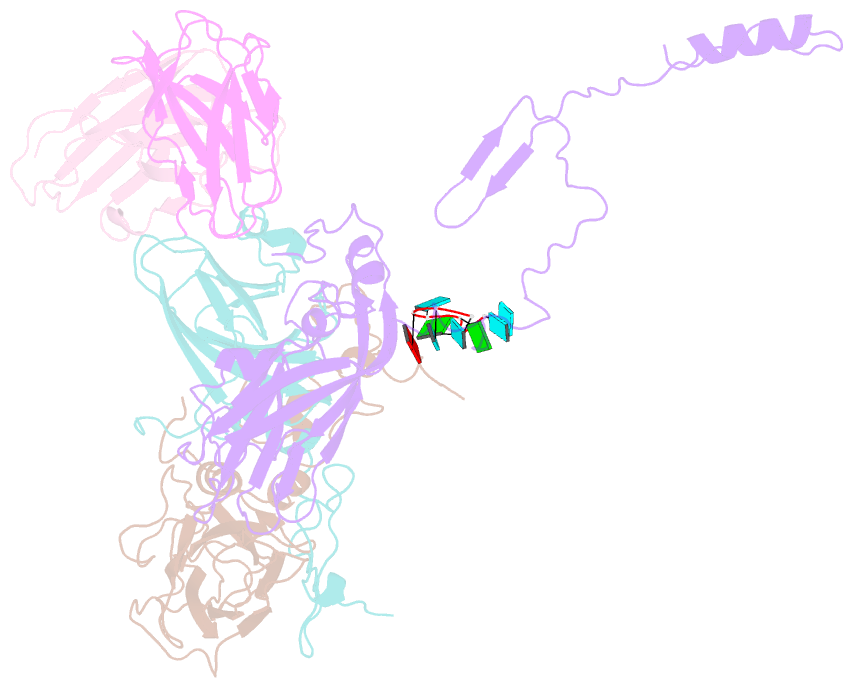
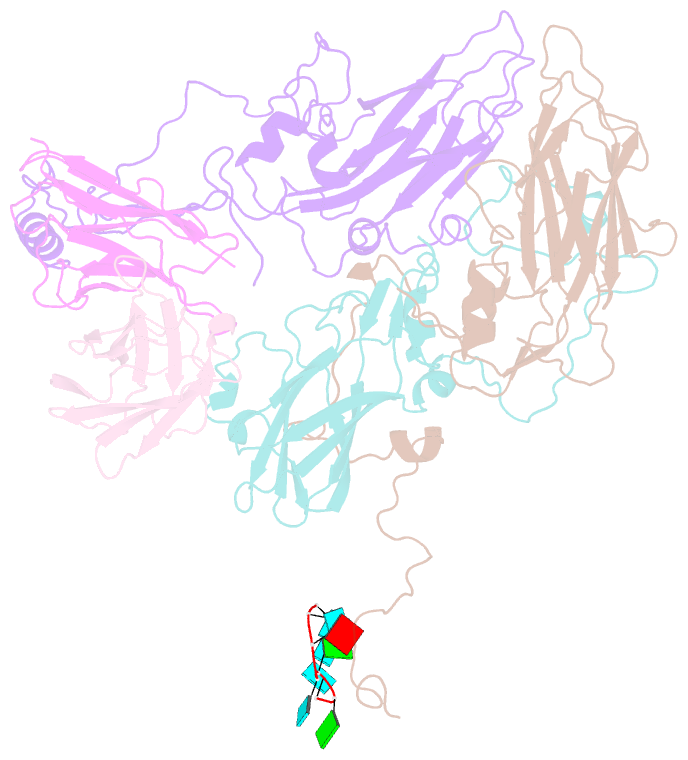
Summary information and primary citation
- PDB-id
- 6gv4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- virus
- Method
- cryo-EM (2.8 Å)
- Summary
- High-resolution cryo-EM of fab-labeled human parechovirus 3
- Reference
- Domanska A, Flatt JW, Jukonen JJJ, Geraets JA, Butcher SJ (2019): "A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody." J.Virol., 93. doi: 10.1128/JVI.01597-18.
- Abstract
- Human parechovirus 3 (HPeV3) infection is associated with sepsis characterized by significant immune activation and subsequent tissue damage in neonates. Strategies to limit infection have been unsuccessful due to inadequate molecular diagnostic tools for early detection and the lack of a vaccine or specific antiviral therapy. Toward the latter, we present a 2.8-Å-resolution structure of HPeV3 in complex with fragments from a neutralizing human monoclonal antibody, AT12-015, using cryo-electron microscopy (cryo-EM) and image reconstruction. Modeling revealed that the epitope extends across neighboring asymmetric units with contributions from capsid proteins VP0, VP1, and VP3. Antibody decoration was found to block binding of HPeV3 to cultured cells. Additionally, at high resolution, it was possible to model a stretch of RNA inside the virion and, from this, identify the key features that drive and stabilize protein-RNA association during assembly.