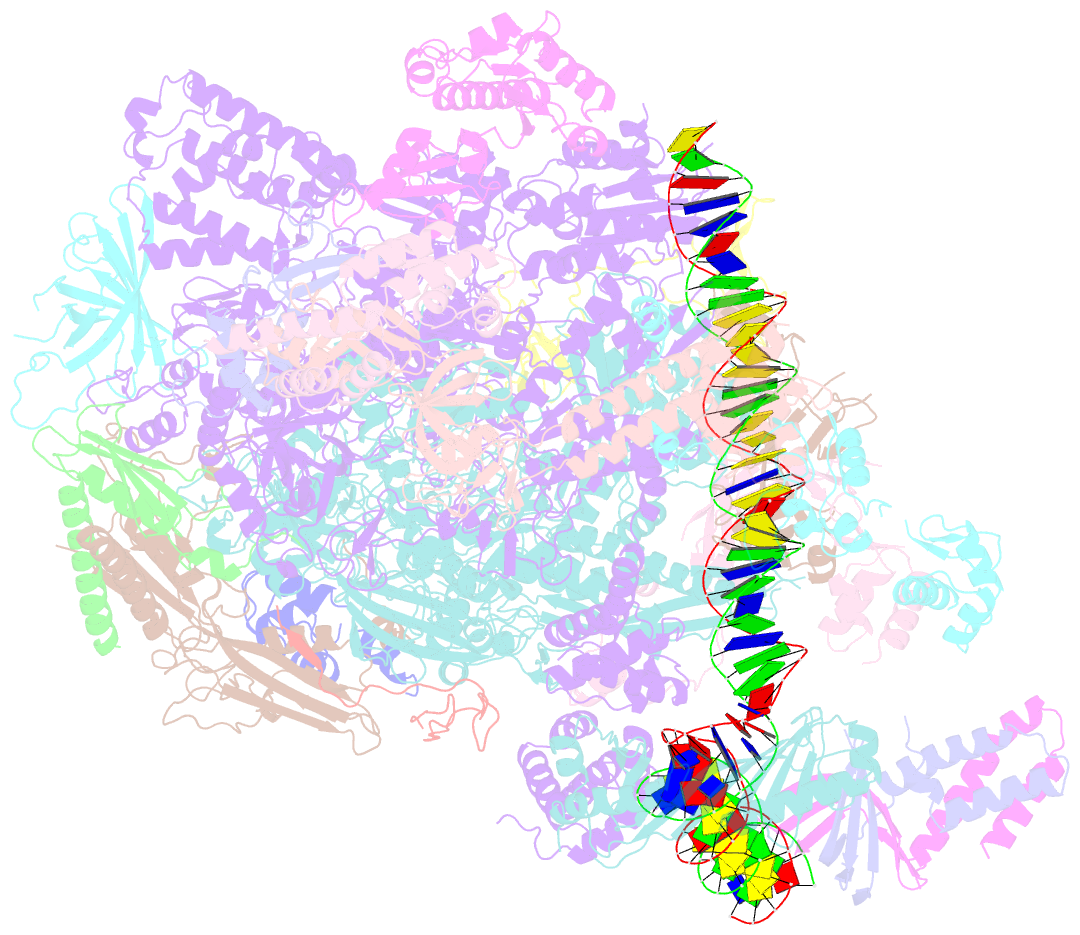
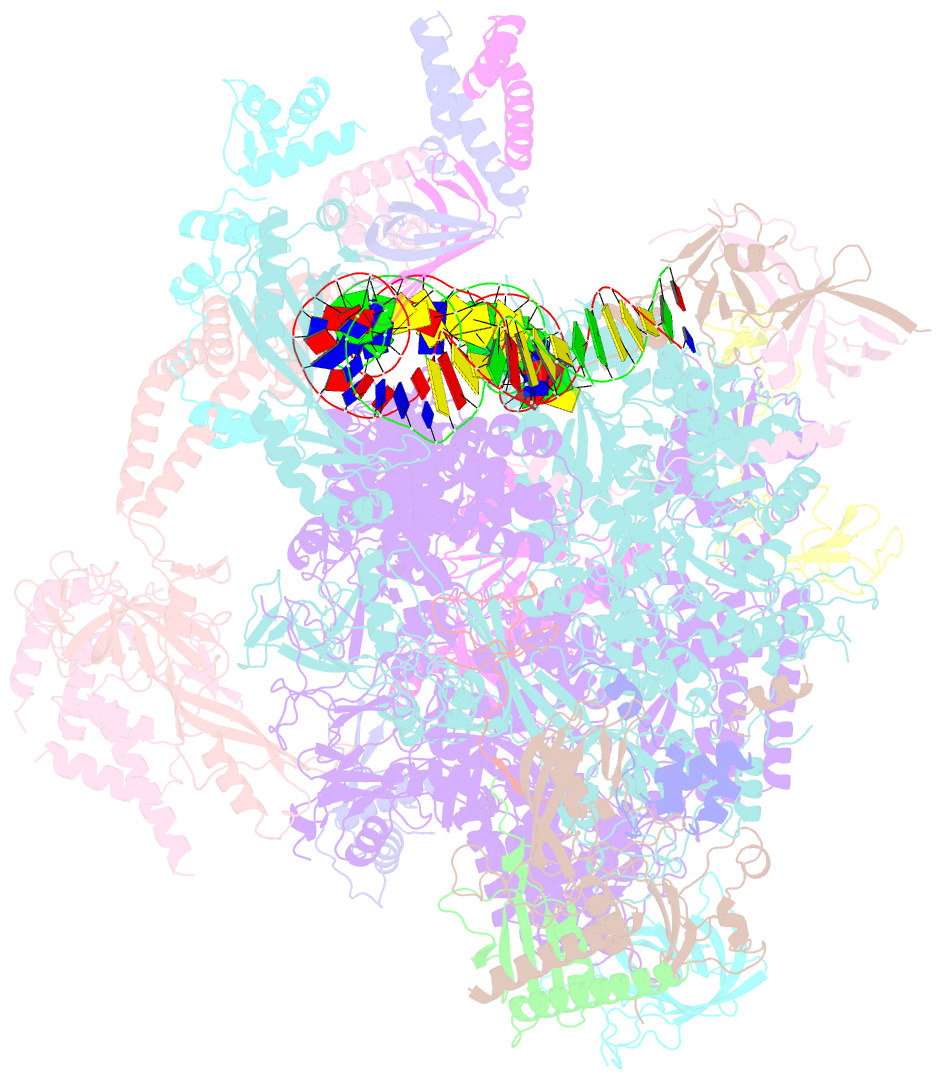
Summary information and primary citation
- PDB-id
- 6gyl; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (4.8 Å)
- Summary
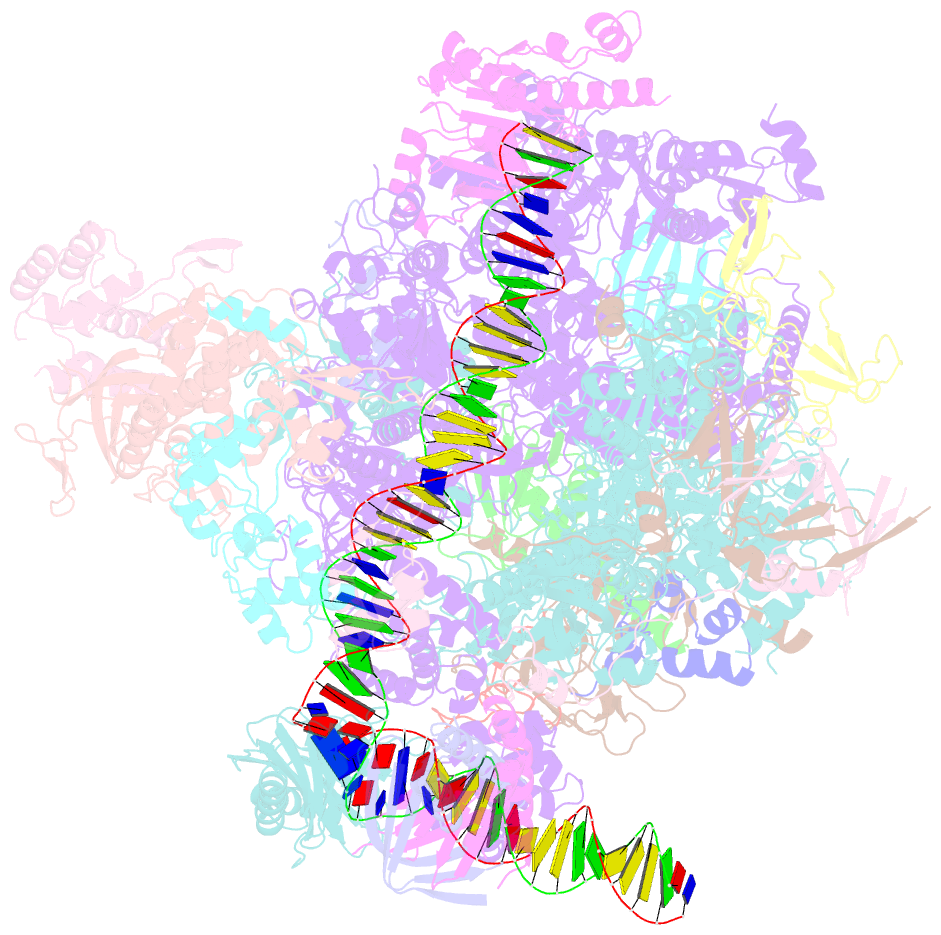
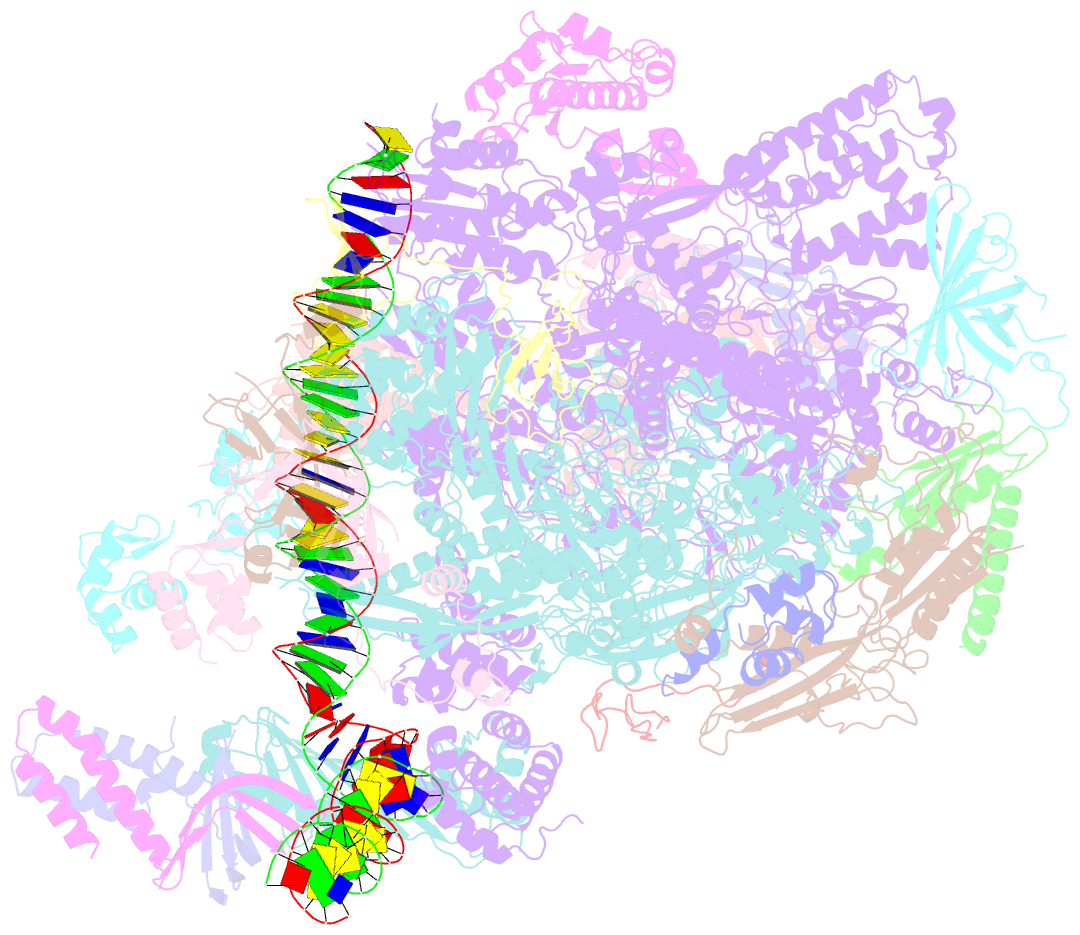
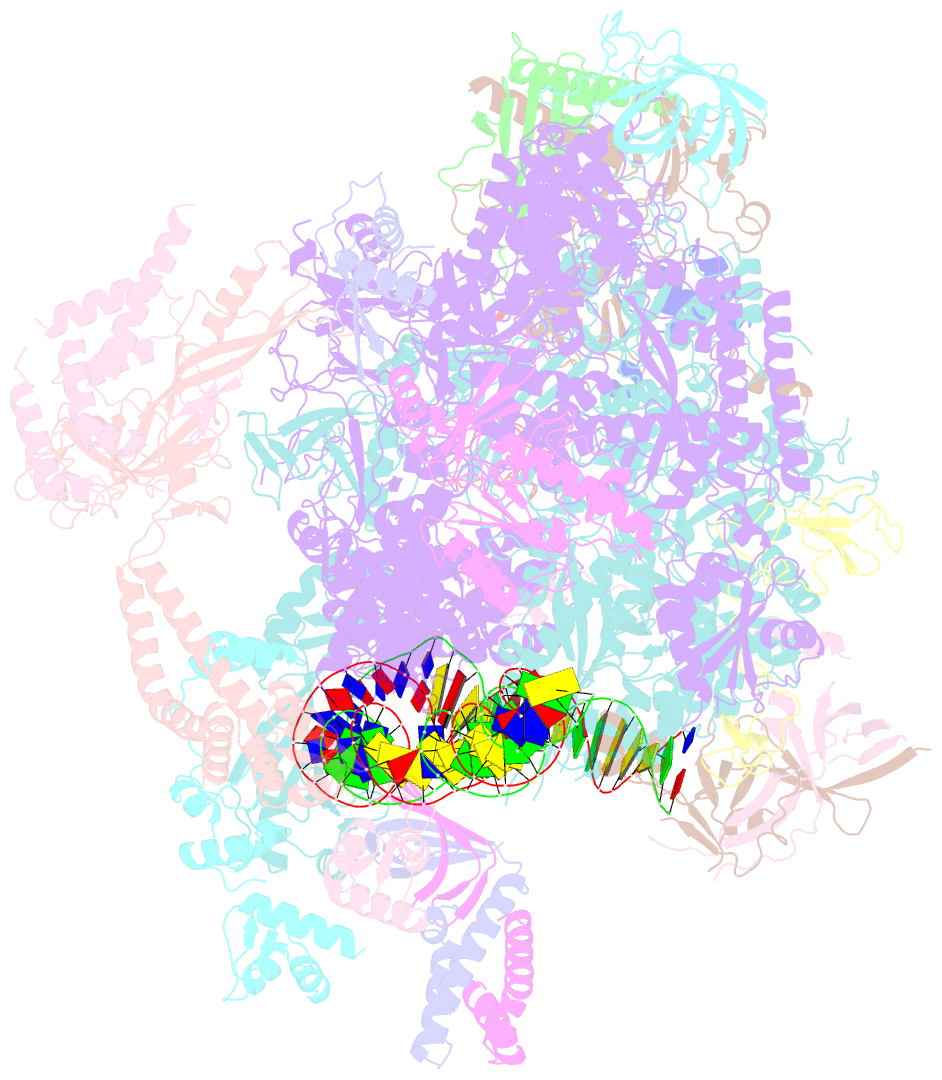
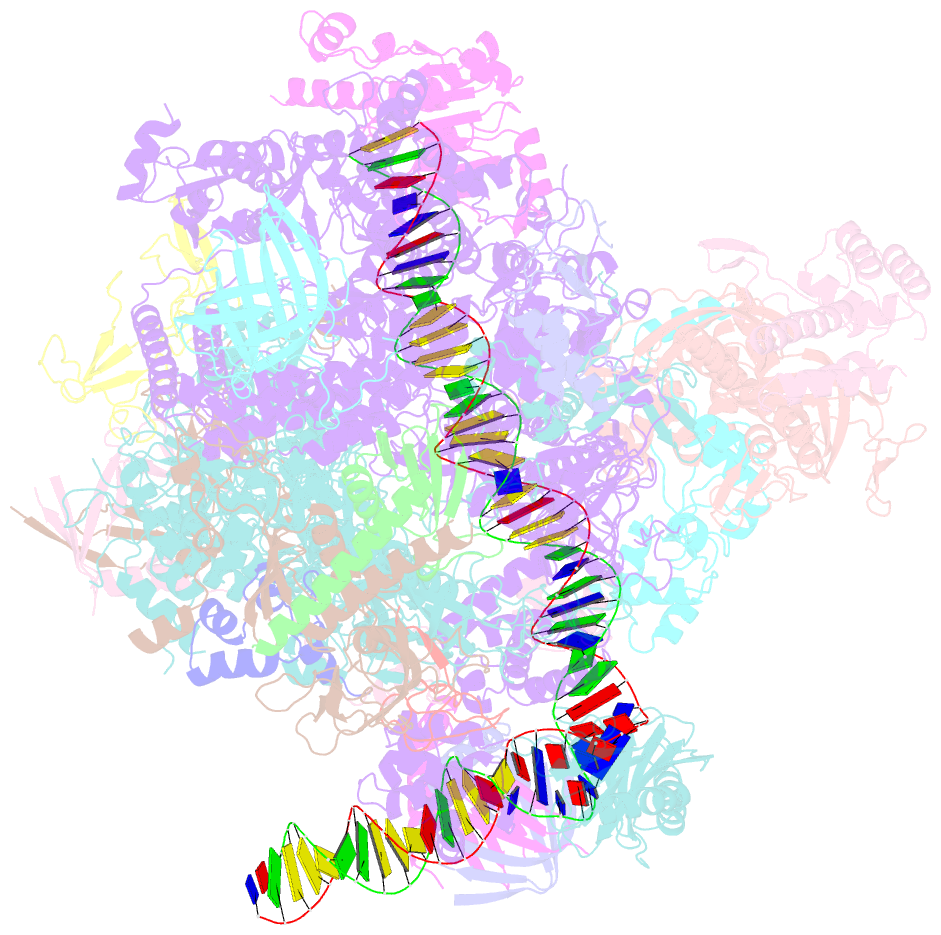
- Structure of a yeast closed complex with distorted DNA (core ccdist)
- Reference
- Dienemann C, Schwalb B, Schilbach S, Cramer P (2019): "Promoter Distortion and Opening in the RNA Polymerase II Cleft." Mol. Cell, 73, 97-106.e4. doi: 10.1016/j.molcel.2018.10.014.
- Abstract
- Transcription initiation requires opening of promoter DNA in the RNA polymerase II (Pol II) pre-initiation complex (PIC), but it remains unclear how this is achieved. Here we report the cryo-electron microscopic (cryo-EM) structure of a yeast PIC that contains underwound, distorted promoter DNA in the closed Pol II cleft. The DNA duplex axis is offset at the upstream edge of the initially melted DNA region (IMR) where DNA opening begins. Unstable IMRs are found in a subset of yeast promoters that we show can still initiate transcription after depletion of the transcription factor (TF) IIH (TFIIH) translocase Ssl2 (XPB in human) from the nucleus in vivo. PIC-induced DNA distortions may thus prime the IMR for melting and may explain how unstable IMRs that are predicted in promoters of Pol I and Pol III can open spontaneously. These results suggest that DNA distortion in the polymerase cleft is a general mechanism that contributes to promoter opening.