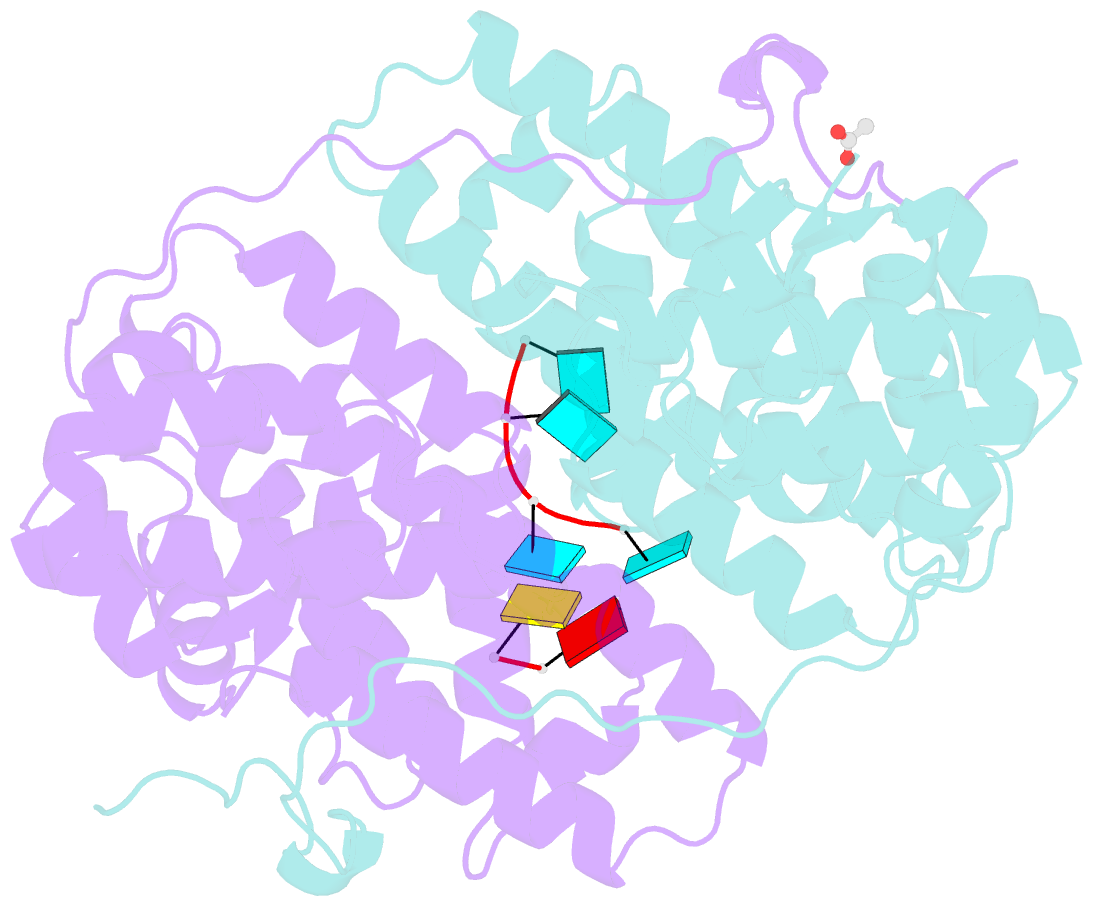
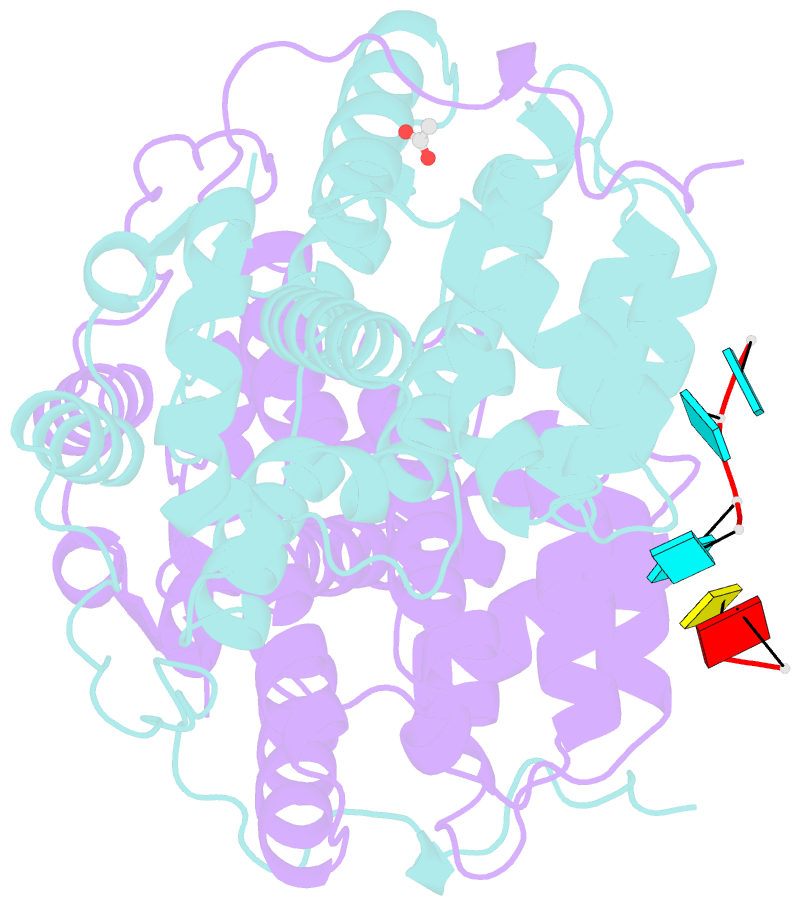
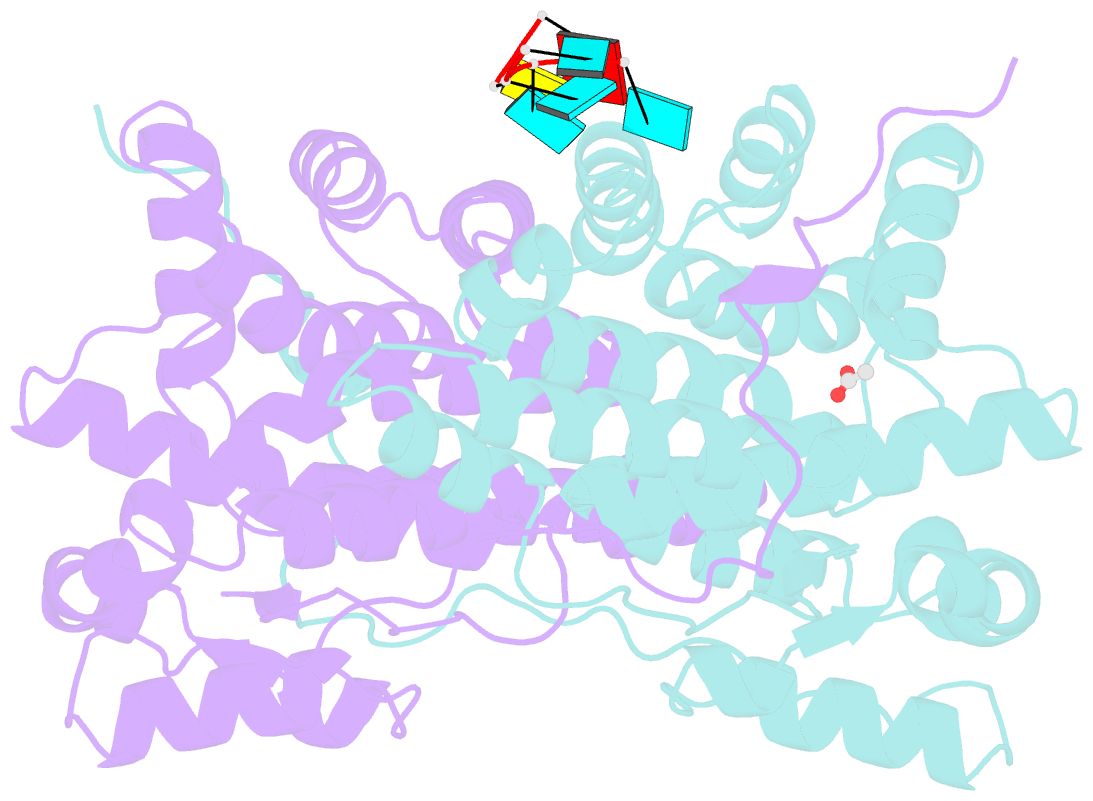
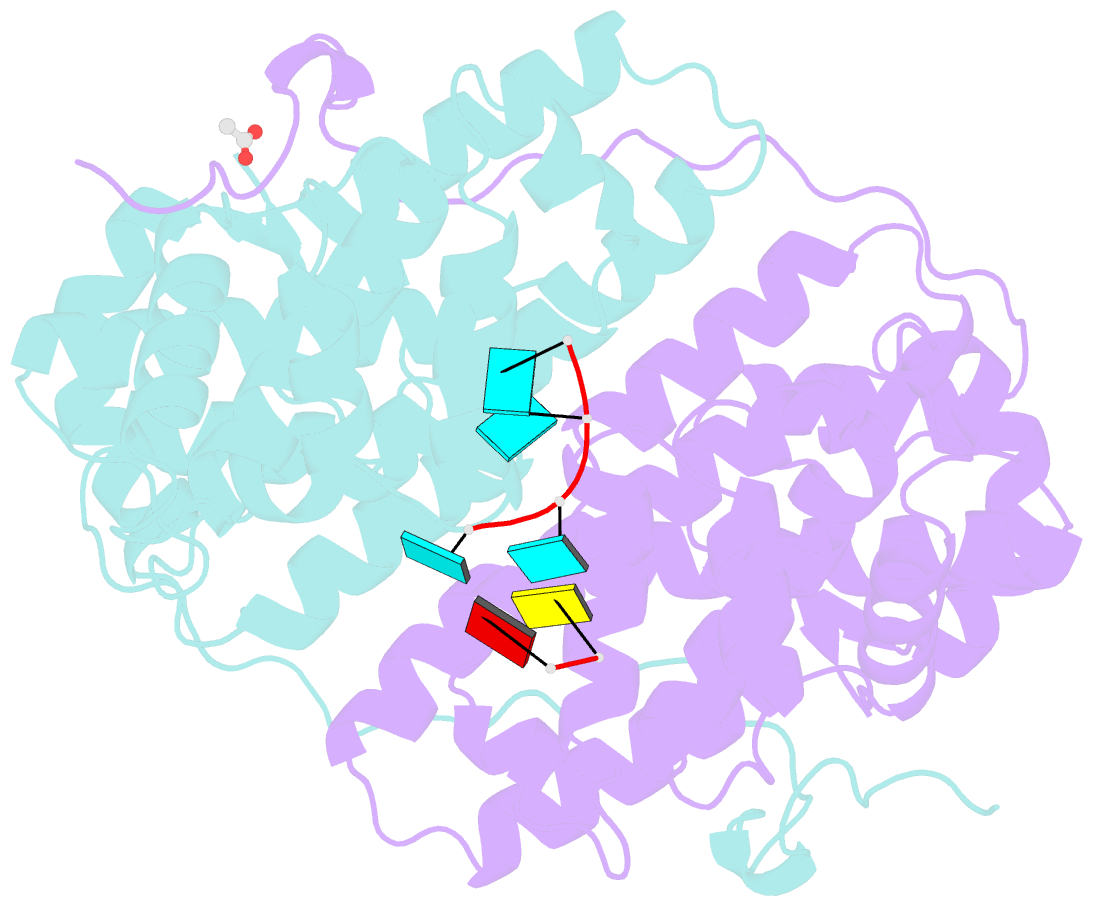
Summary information and primary citation
- PDB-id
- 6hau; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- X-ray (1.86 Å)
- Summary
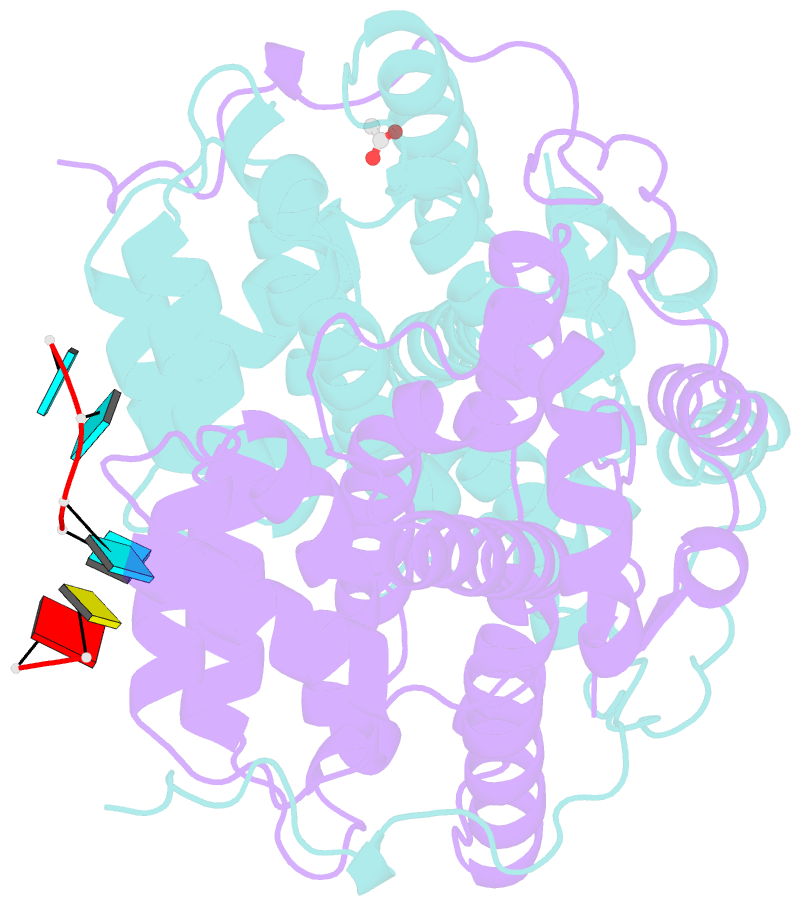
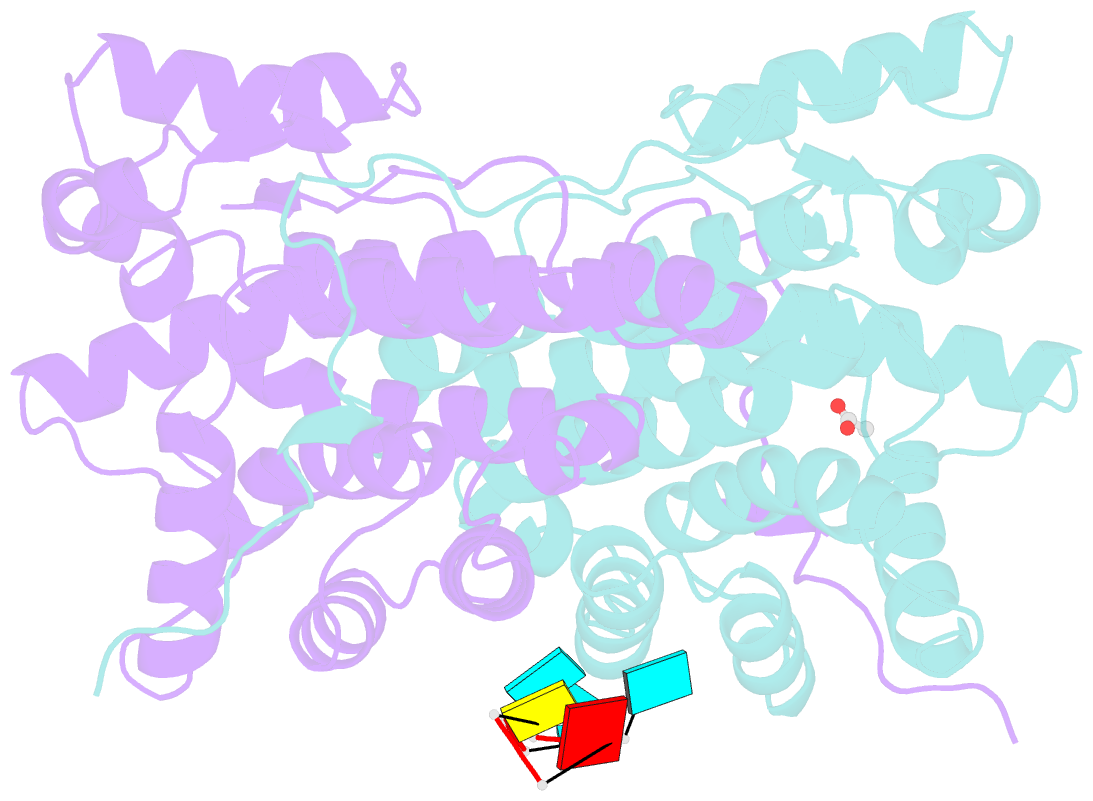
- Kshv pan RNA mta-response element fragment complexed with the globular domain of herpesvirus saimiri orf57
- Reference
- Tunnicliffe RB, Levy C, Ruiz Nivia HD, Sandri-Goldin RM, Golovanov AP (2019): "Structural identification of conserved RNA binding sites in herpesvirus ORF57 homologs: implications for PAN RNA recognition." Nucleic Acids Res., 47, 1987-2001. doi: 10.1093/nar/gky1181.
- Abstract
- Kaposi's sarcoma-associated herpesvirus (KSHV) transcribes a long noncoding polyadenylated nuclear (PAN) RNA, which promotes the latent to lytic transition by repressing host genes involved in antiviral responses as well as viral proteins that support the latent state. KSHV also expresses several early proteins including ORF57 (Mta), a member of the conserved multifunctional ICP27 protein family, which is essential for productive replication. ORF57/Mta interacts with PAN RNA via a region termed the Mta responsive element (MRE), stabilizing the transcript and supporting nuclear accumulation. Here, using a close homolog of KSHV ORF57 from herpesvirus saimiri (HVS), we determined the crystal structure of the globular domain in complex with a PAN RNA MRE, revealing a uracil specific binding site that is also conserved in KSHV. Using solution NMR, RNA binding was also mapped within the disordered N-terminal domain of KSHV ORF57, and showed specificity for an RNA fragment containing a GAAGRG motif previously known to bind a homologous region in HVS ORF57. Together these data located novel differential RNA recognition sites within neighboring domains of herpesvirus ORF57 homologs, and revealed high-resolution details of their interactions with PAN RNA, thus providing insight into interactions crucial to viral function.