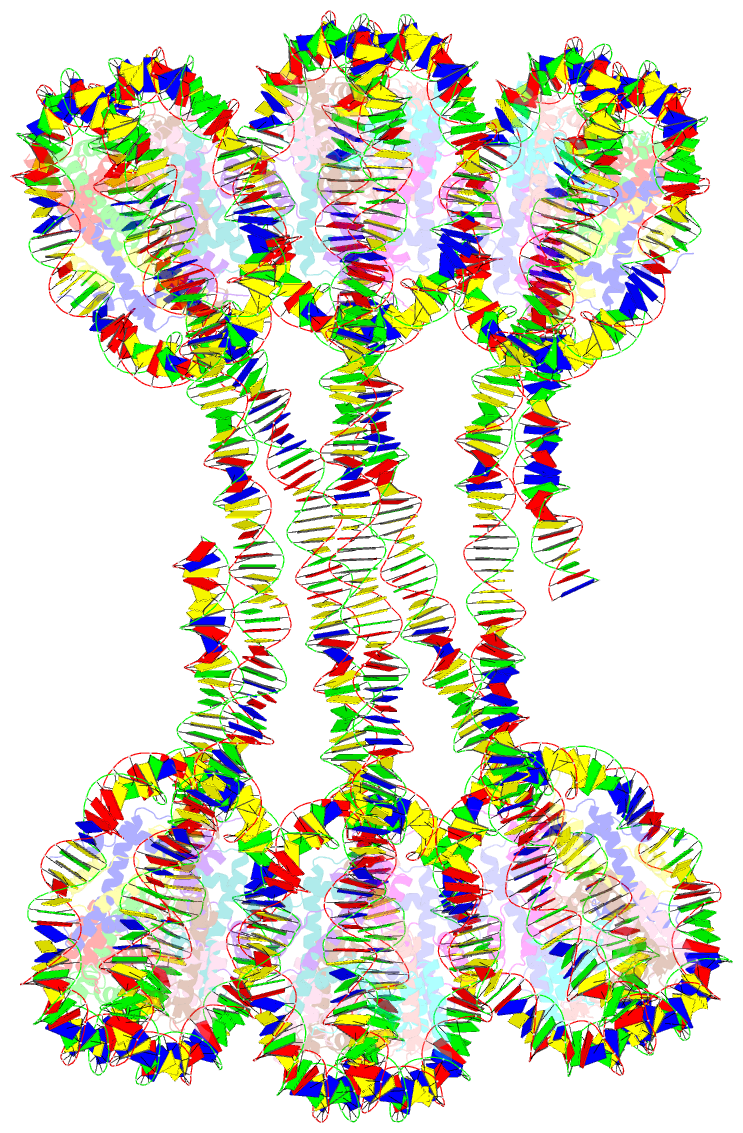
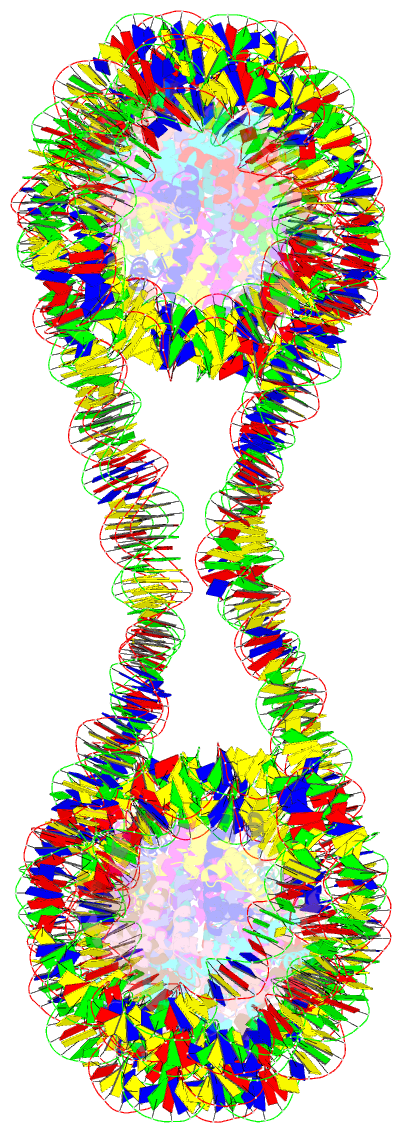
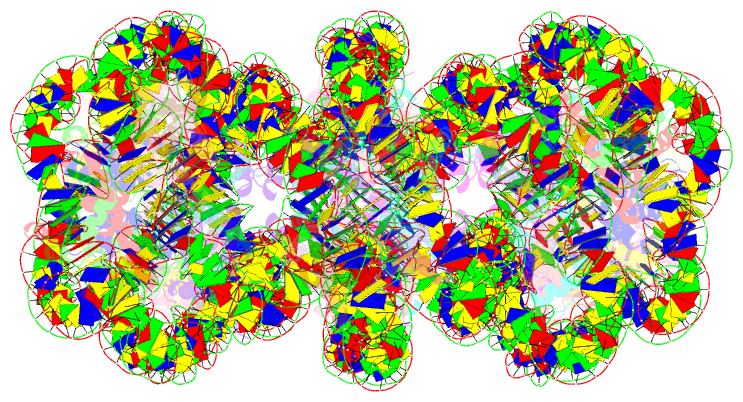
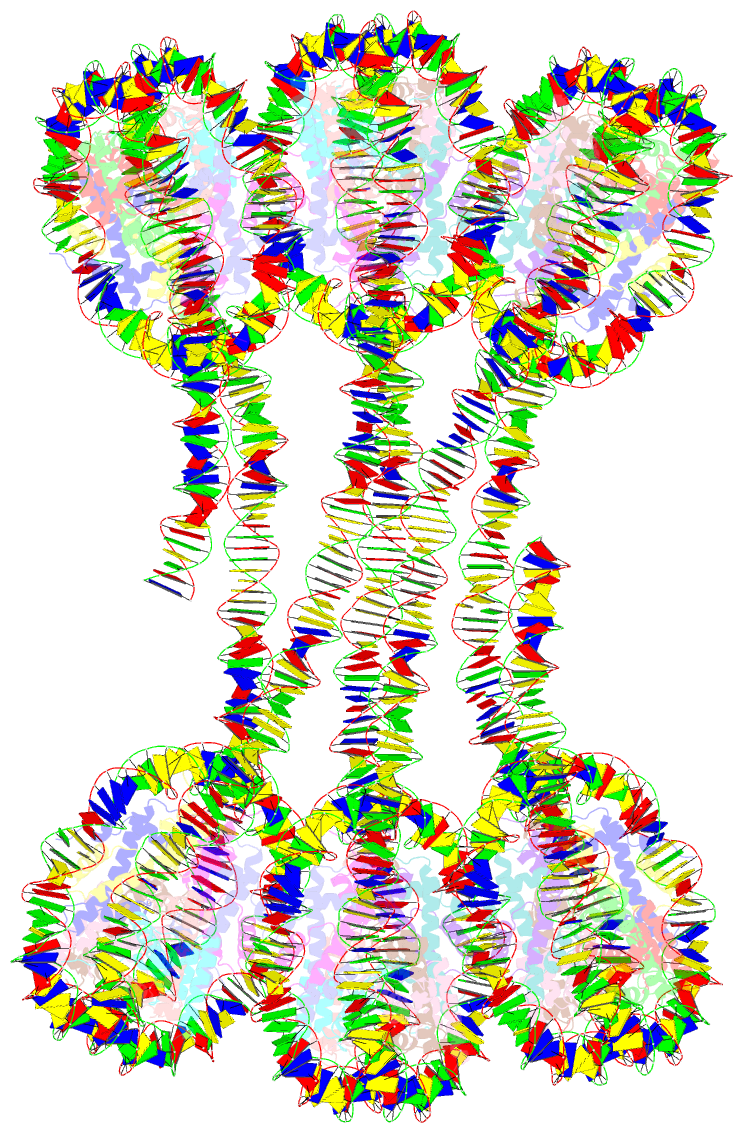
Summary information and primary citation
- PDB-id
- 6hkt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (9.7 Å)
- Summary
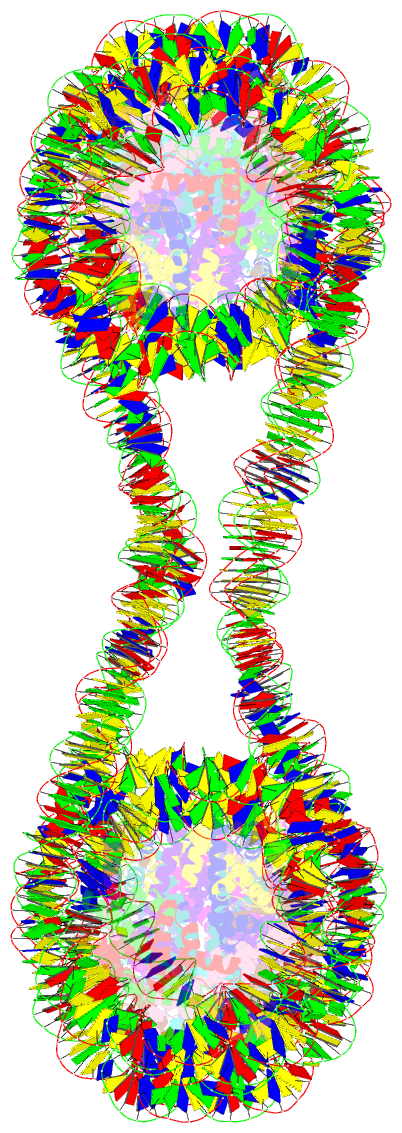
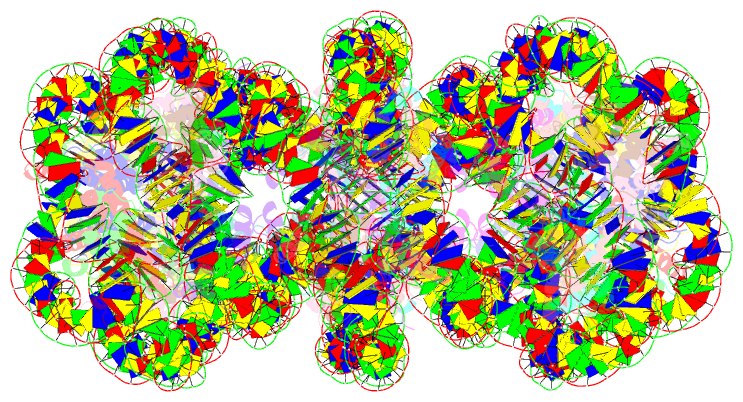
- Structure of an h1-bound 6-nucleosome array
- Reference
- Garcia-Saez I, Menoni H, Boopathi R, Shukla MS, Soueidan L, Noirclerc-Savoye M, Le Roy A, Skoufias DA, Bednar J, Hamiche A, Angelov D, Petosa C, Dimitrov S (2018): "Structure of an H1-Bound 6-Nucleosome Array Reveals an Untwisted Two-Start Chromatin Fiber Conformation." Mol. Cell, 72, 902-915.e7. doi: 10.1016/j.molcel.2018.09.027.
- Abstract
- Chromatin adopts a diversity of regular and irregular fiber structures in vitro and in vivo. However, how an array of nucleosomes folds into and switches between different fiber conformations is poorly understood. We report the 9.7 Å resolution crystal structure of a 6-nucleosome array bound to linker histone H1 determined under ionic conditions that favor incomplete chromatin condensation. The structure reveals a flat two-start helix with uniform nucleosomal stacking interfaces and a nucleosome packing density that is only half that of a twisted 30-nm fiber. Hydroxyl radical footprinting indicates that H1 binds the array in an on-dyad configuration resembling that observed for mononucleosomes. Biophysical, cryo-EM, and crosslinking data validate the crystal structure and reveal that a minor change in ionic environment shifts the conformational landscape to a more compact, twisted form. These findings provide insights into the structural plasticity of chromatin and suggest a possible assembly pathway for a 30-nm fiber.