Summary information and primary citation
- PDB-id
- 6i52; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (4.7 Å)
- Summary
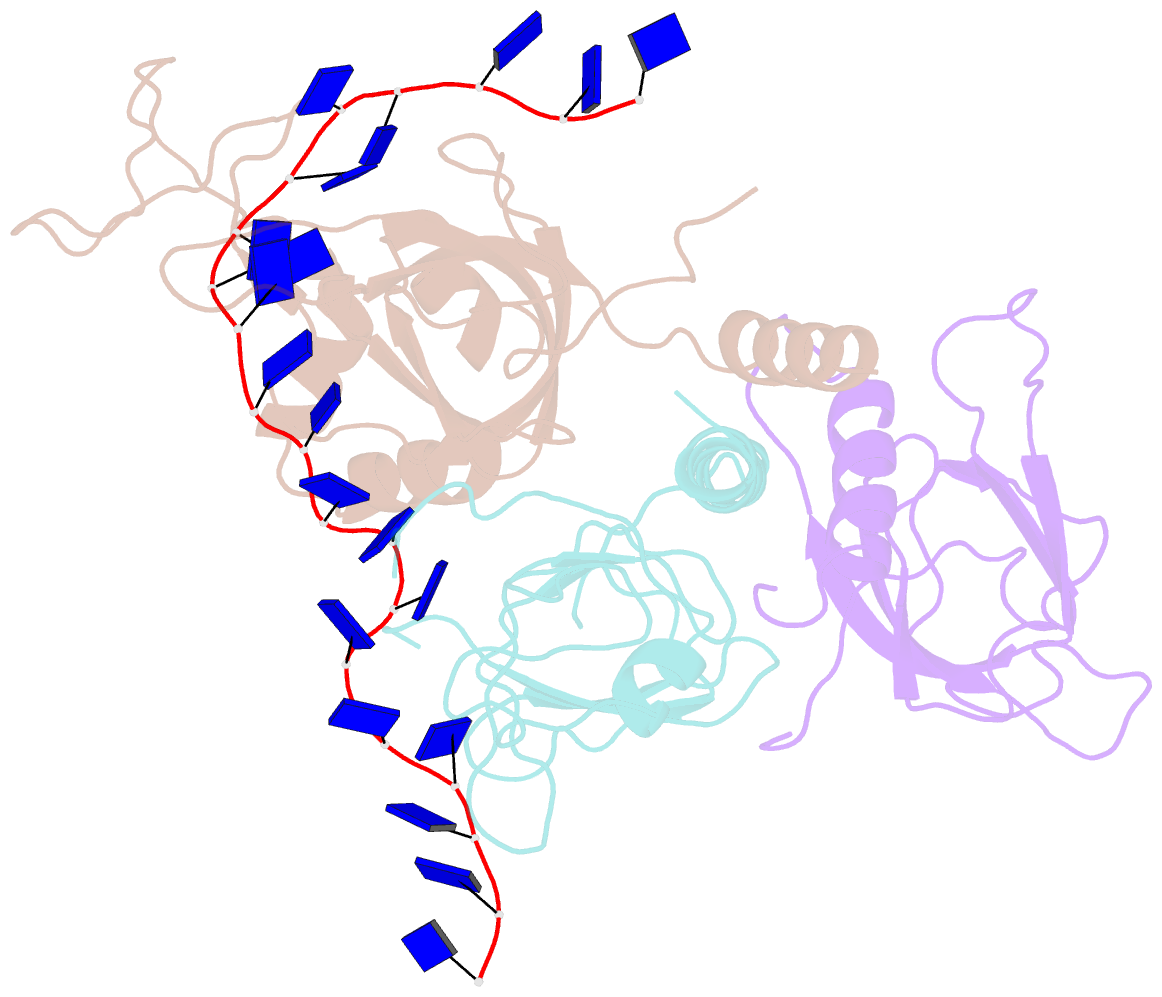
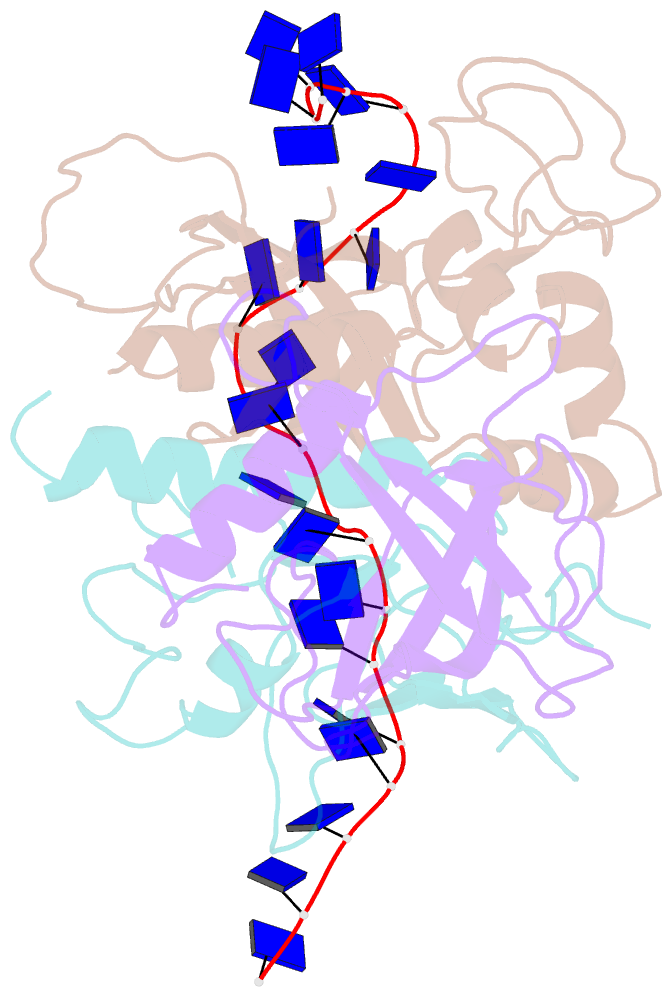
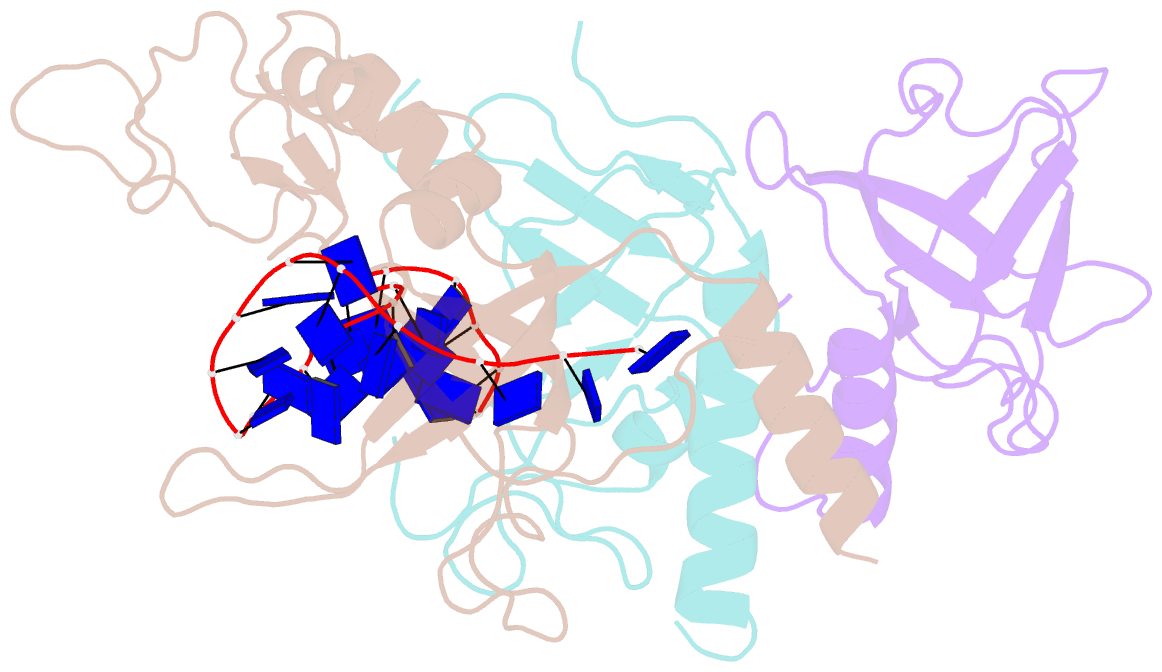
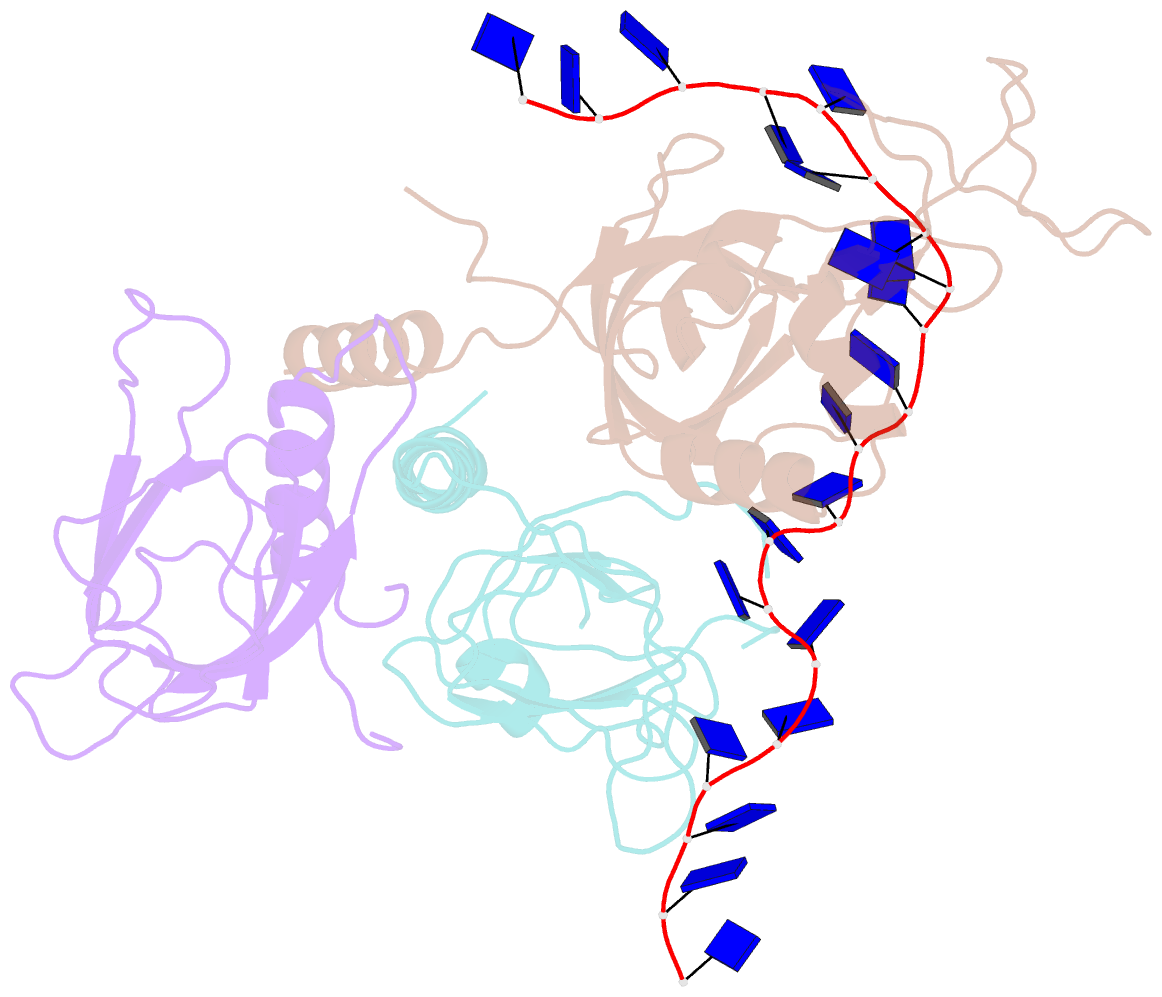
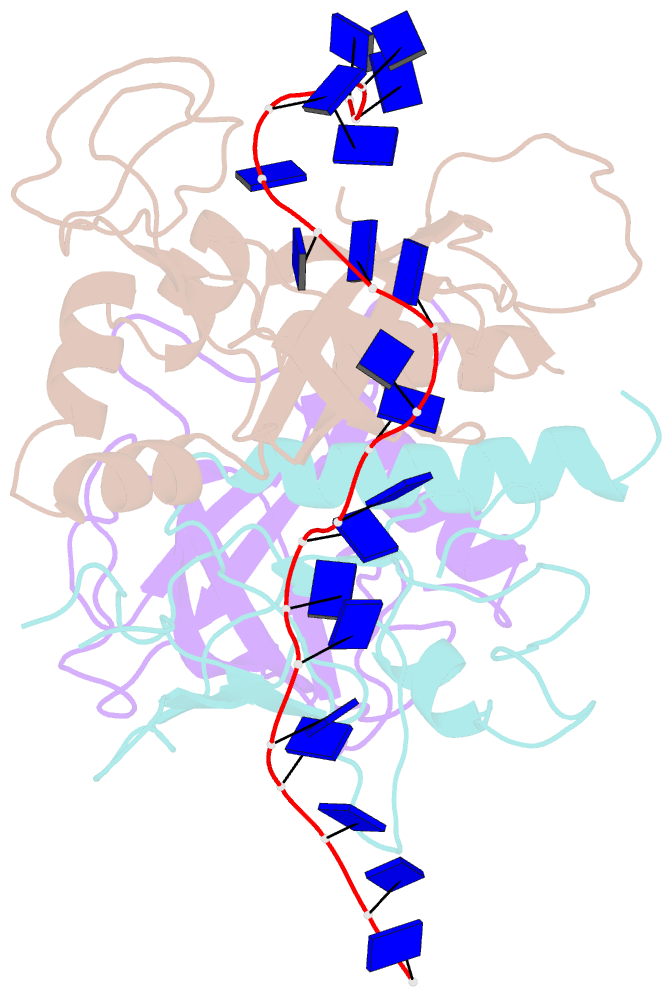
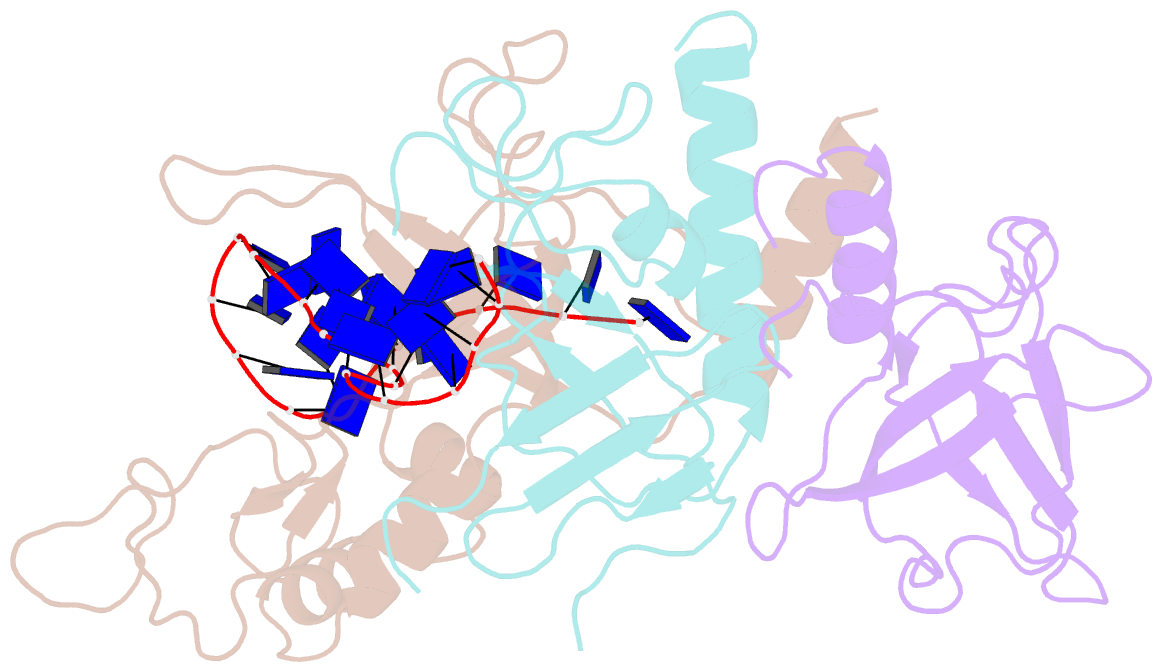
- Yeast rpa bound to ssDNA
- Reference
- Yates LA, Aramayo RJ, Pokhrel N, Caldwell CC, Kaplan JA, Perera RL, Spies M, Antony E, Zhang X (2018): "A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA." Nat Commun, 9, 5447. doi: 10.1038/s41467-018-07883-7.
- Abstract
- Replication Protein A (RPA), the major eukaryotic single stranded DNA-binding protein, binds to exposed ssDNA to protect it from nucleases, participates in a myriad of nucleic acid transactions and coordinates the recruitment of other important players. RPA is a heterotrimer and coats long stretches of single-stranded DNA (ssDNA). The precise molecular architecture of the RPA subunits and its DNA binding domains (DBDs) during assembly is poorly understood. Using cryo electron microscopy we obtained a 3D reconstruction of the RPA trimerisation core bound with ssDNA (∼55 kDa) at ∼4.7 Å resolution and a dimeric RPA assembly on ssDNA. FRET-based solution studies reveal dynamic rearrangements of DBDs during coordinated RPA binding and this activity is regulated by phosphorylation at S178 in RPA70. We present a structural model on how dynamic DBDs promote the cooperative assembly of multiple RPAs on long ssDNA.