Summary information and primary citation
- PDB-id
- 6iuc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.4 Å)
- Summary
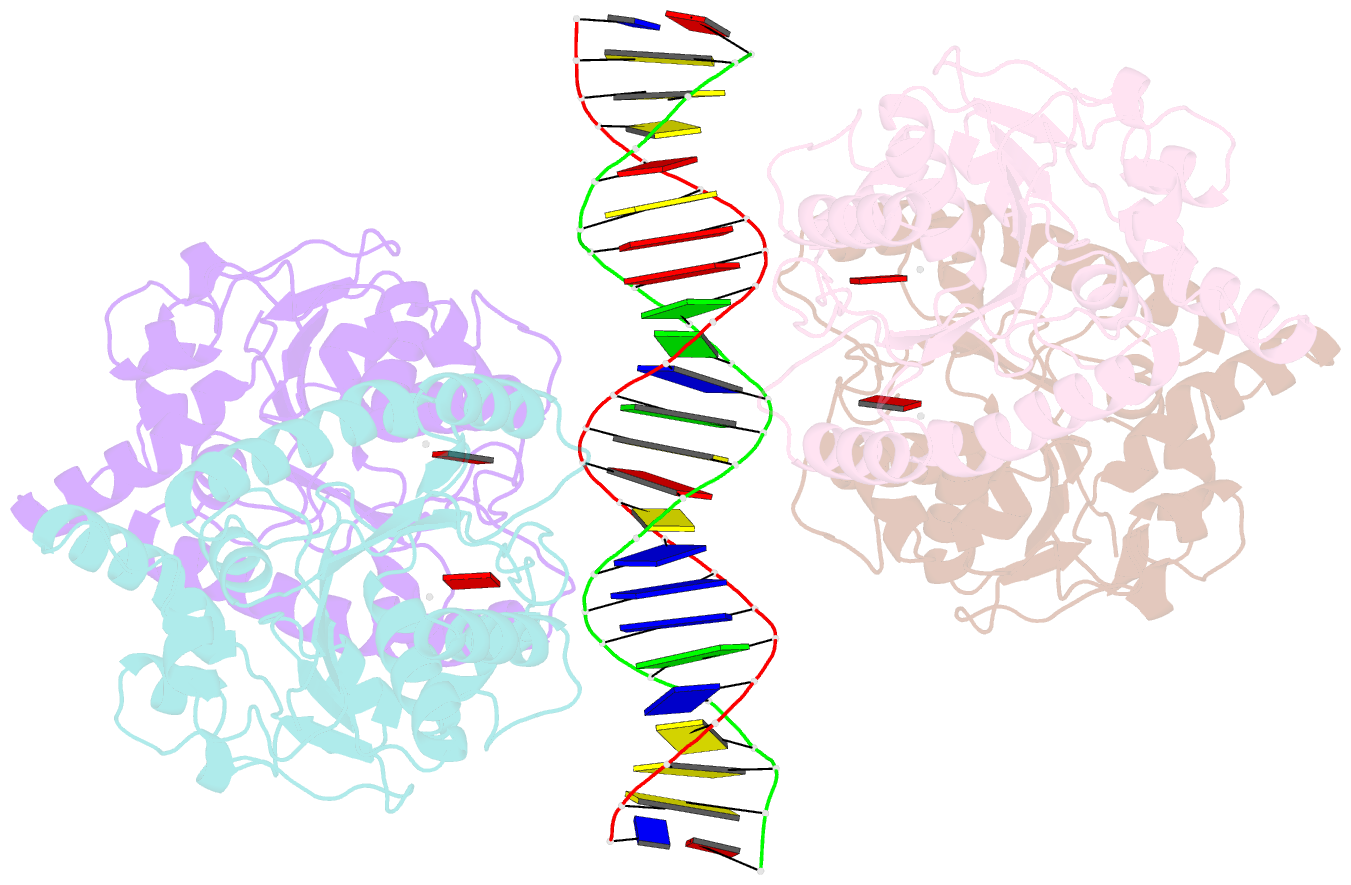
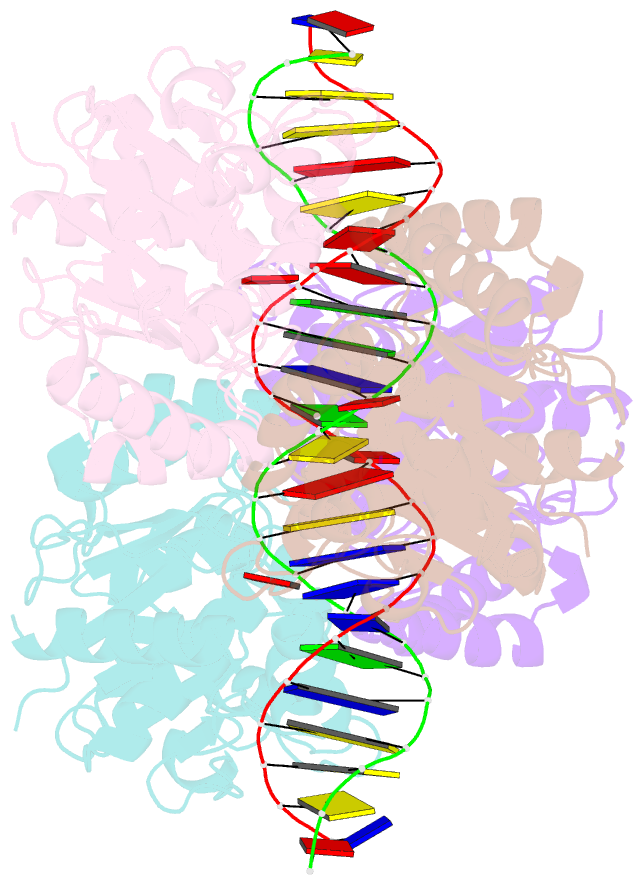
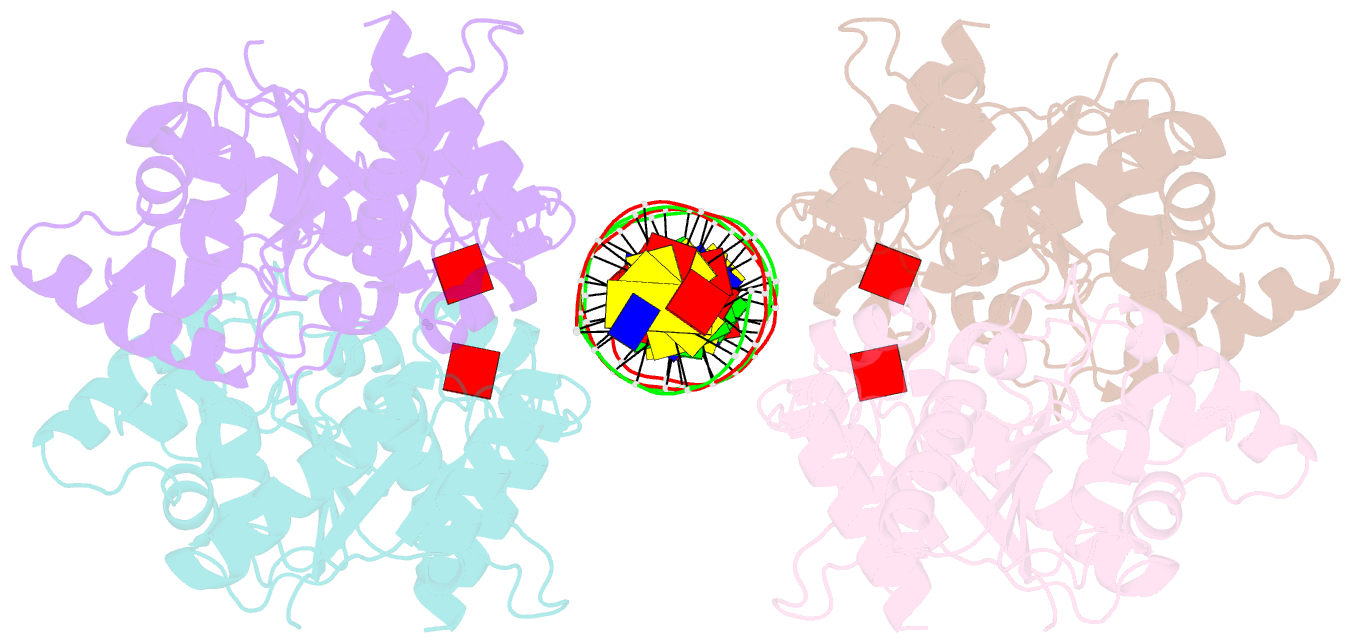
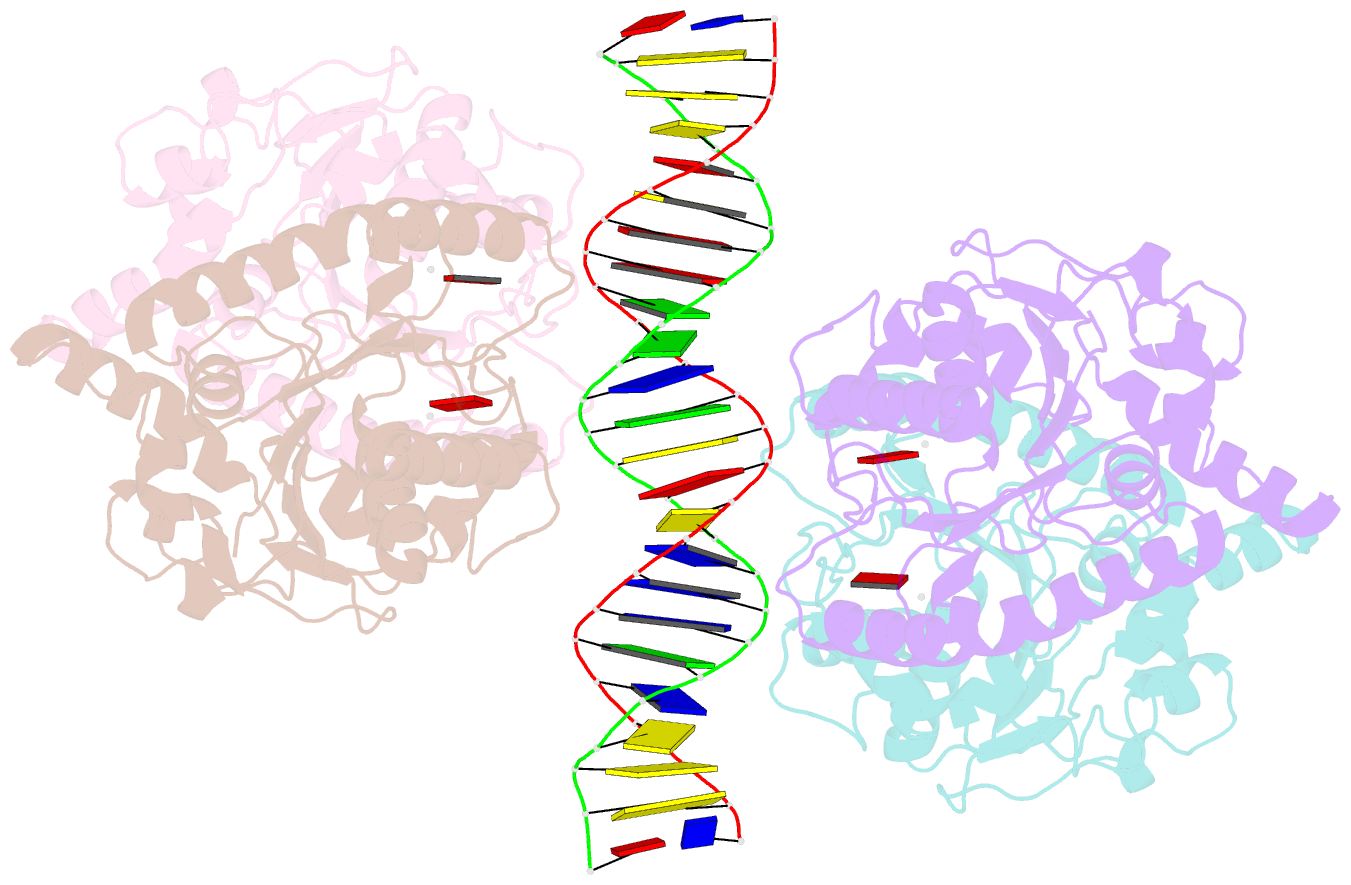
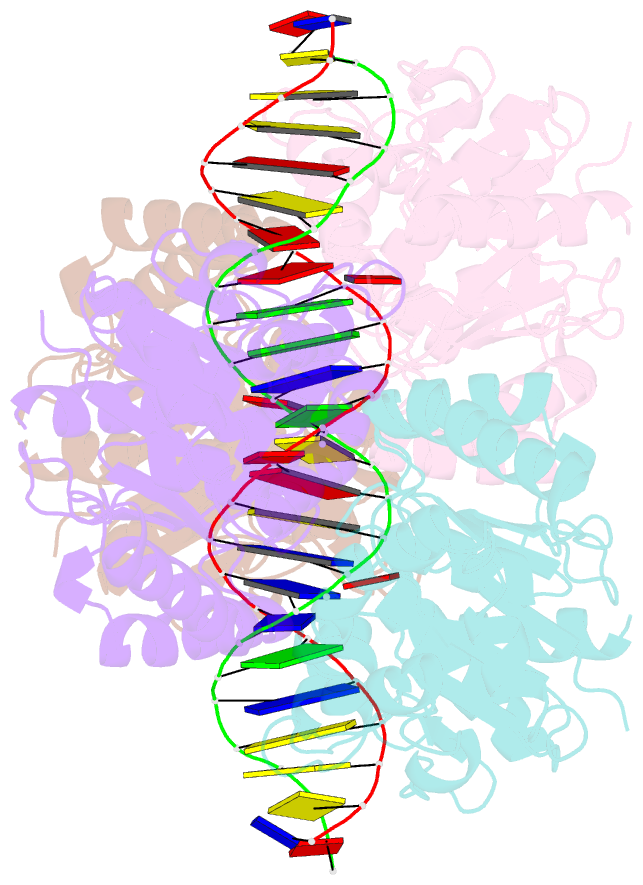
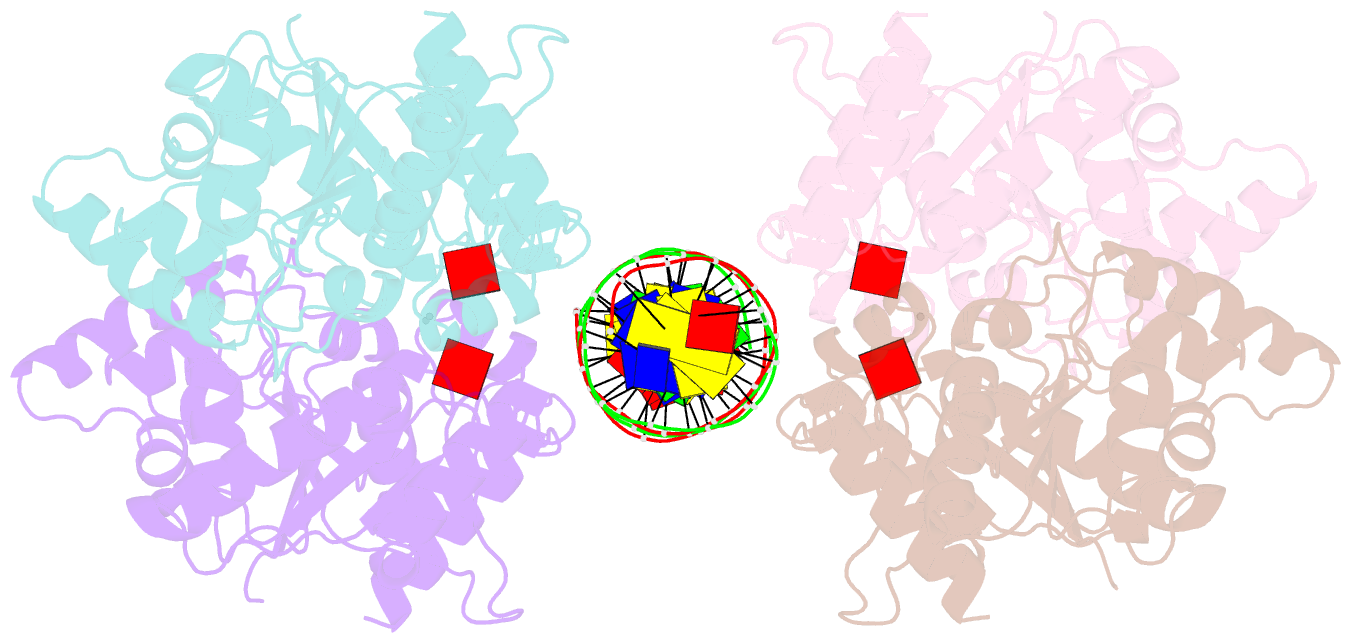
- Structure of helicobacter pylori soj-atp complex bound to DNA
- Reference
- Chu CH, Yen CY, Chen BW, Lin MG, Wang LH, Tang KZ, Hsiao CD, Sun YJ (2019): "Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation." Nucleic Acids Res., 47, 2113-2129. doi: 10.1093/nar/gky1251.
- Abstract
- ParABS, an important DNA partitioning process in chromosome segregation, includes ParA (an ATPase), ParB (a parS binding protein) and parS (a centromere-like DNA). The homologous proteins of ParA and ParB in Helicobacter pylori are HpSoj and HpSpo0J, respectively. We analyzed the ATPase activity of HpSoj and found that it is enhanced by both DNA and HpSpo0J. Crystal structures of HpSoj and its DNA complexes revealed a typical ATPase fold and that it is dimeric. DNA binding by HpSoj is promoted by ATP. The HpSoj-ATP-DNA complex non-specifically binds DNA through a continuous basic binding patch formed by lysine residues, with a single DNA-binding site. This complex exhibits a DNA-binding adept state with an active ATP-bound conformation, whereas the HpSoj-ADP-DNA complex may represent a transient DNA-bound state. Based on structural comparisons, HpSoj exhibits a similar DNA binding surface to the bacterial ParA superfamily, but the archaeal ParA superfamily exhibits distinct non-specific DNA-binding via two DNA-binding sites. We detected the HpSpo0J-HpSoj-DNA complex by electron microscopy and show that this nucleoid-adaptor complex (NAC) is formed through HpSoj and HpSpo0J interaction and parS DNA binding. NAC formation is promoted by HpSoj participation and specific parS DNA facilitation.