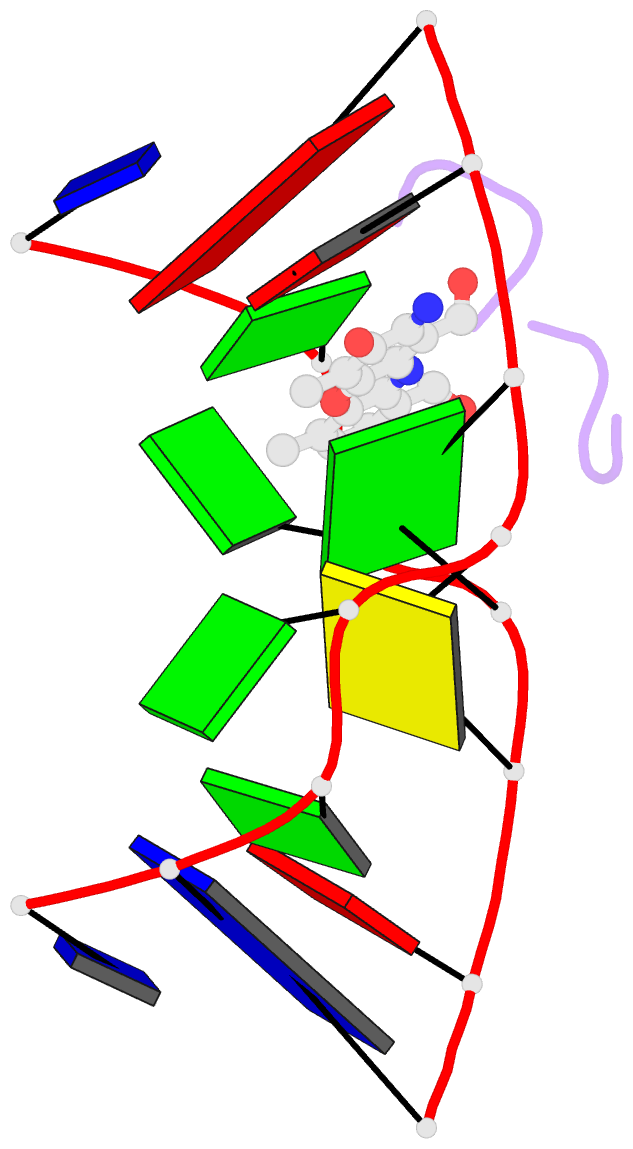
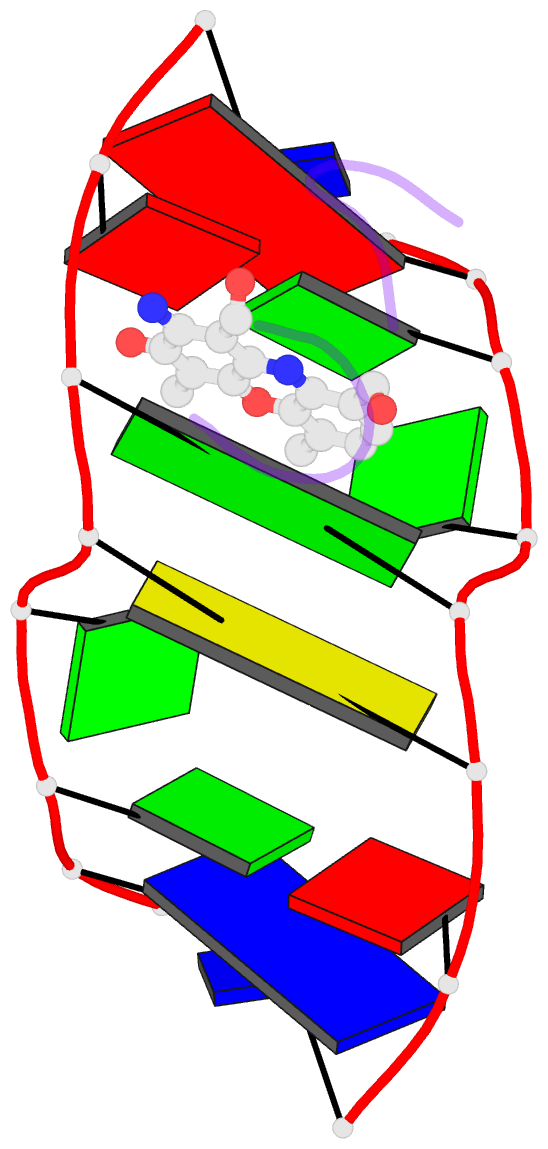
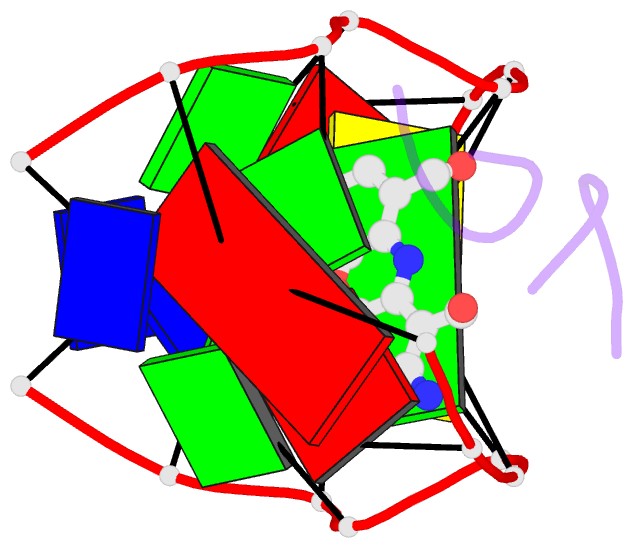
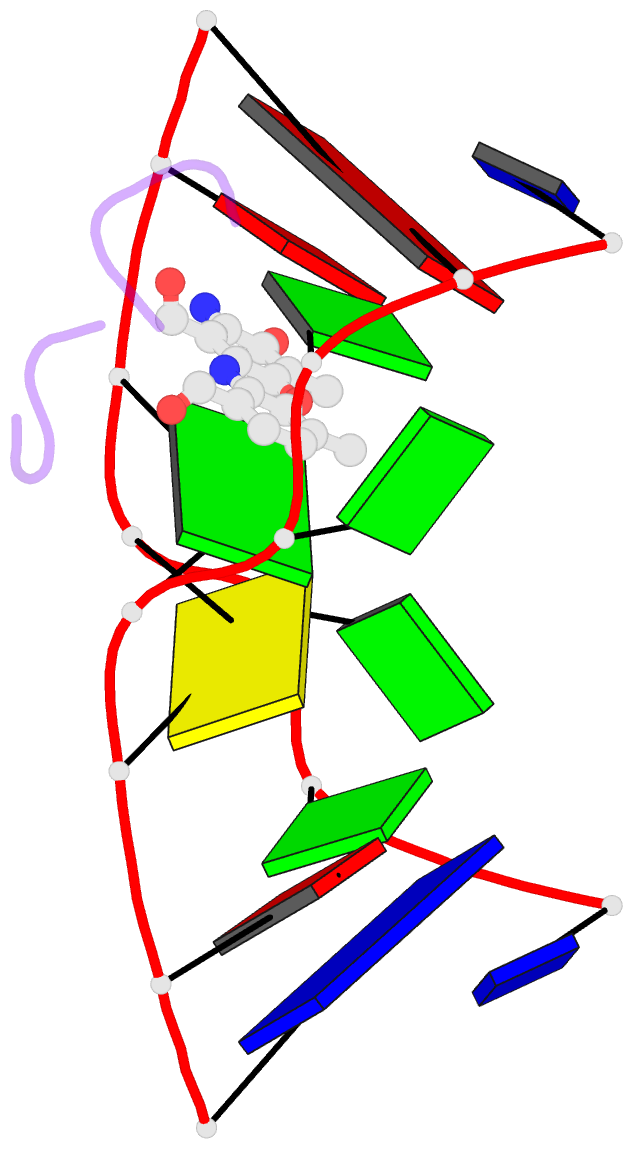
Summary information and primary citation
- PDB-id
- 6j0h; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA-antibiotic
- Method
- X-ray (1.52 Å)
- Summary
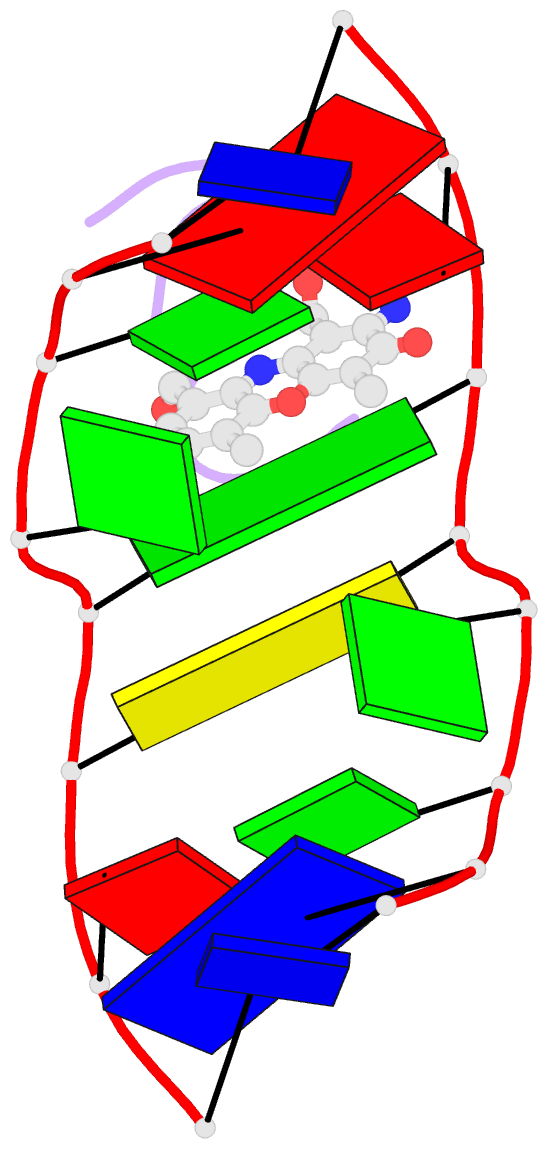
- Crystal structure of actinomycin d- d(ttggcgaa) complex
- Reference
- Satange R, Chuang CY, Neidle S, Hou MH (2019): "Polymorphic G:G mismatches act as hotspots for inducing right-handed Z DNA by DNA intercalation." Nucleic Acids Res., 47, 8899-8912. doi: 10.1093/nar/gkz653.
- Abstract
- DNA mismatches are highly polymorphic and dynamic in nature, albeit poorly characterized structurally. We utilized the antitumour antibiotic CoII(Chro)2 (Chro = chromomycin A3) to stabilize the palindromic duplex d(TTGGCGAA) DNA with two G:G mismatches, allowing X-ray crystallography-based monitoring of mismatch polymorphism. For the first time, the unusual geometry of several G:G mismatches including syn-syn, water mediated anti-syn and syn-syn-like conformations can be simultaneously observed in the crystal structure. The G:G mismatch sites of the d(TTGGCGAA) duplex can also act as a hotspot for the formation of alternative DNA structures with a GC/GA-5' intercalation site for binding by the GC-selective intercalator actinomycin D (ActiD). Direct intercalation of two ActiD molecules to G:G mismatch sites causes DNA rearrangements, resulting in backbone distortion to form right-handed Z-DNA structures with a single-step sharp kink. Our study provides insights on intercalators-mismatch DNA interactions and a rationale for mismatch interrogation and detection via DNA intercalation.