Summary information and primary citation
- PDB-id
- 6j9e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.41 Å)
- Summary
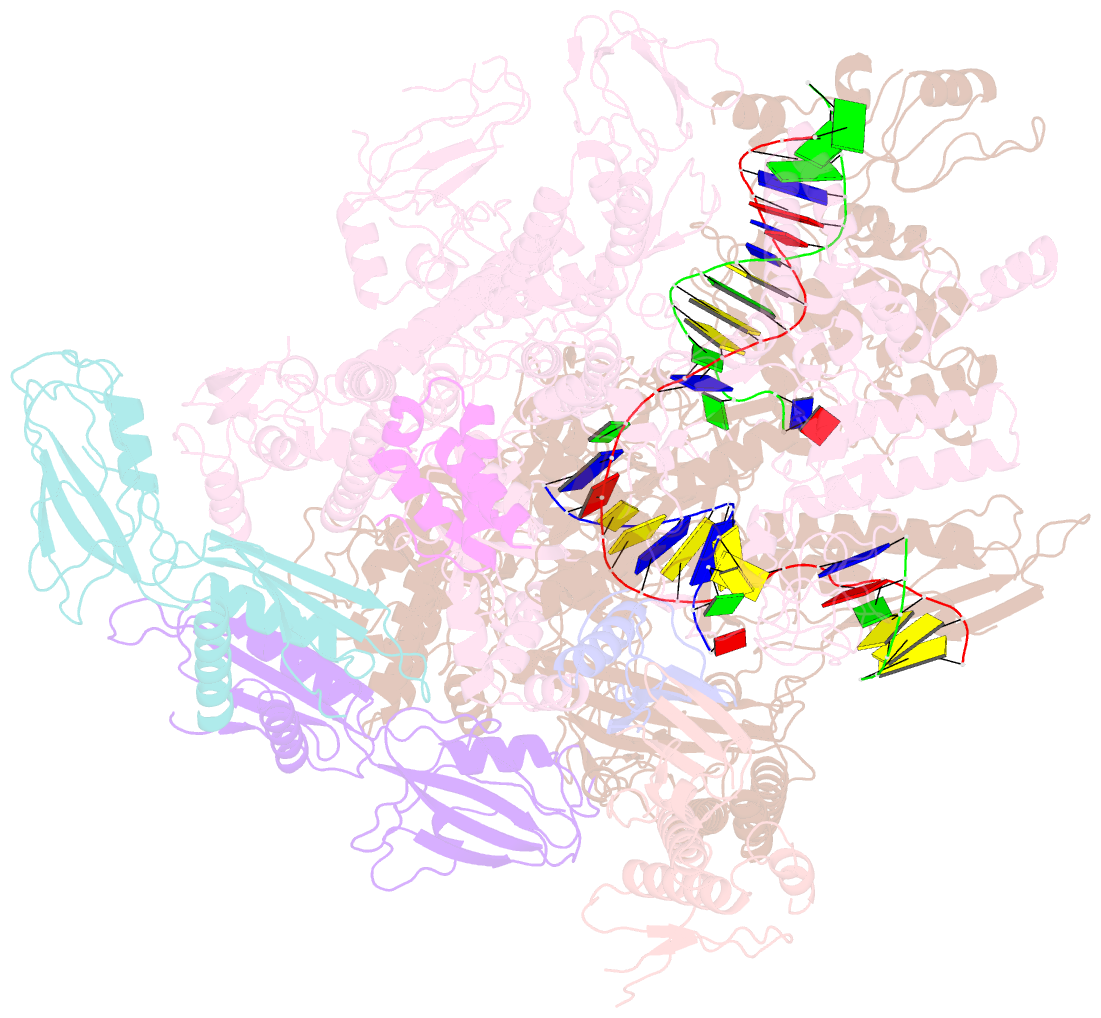
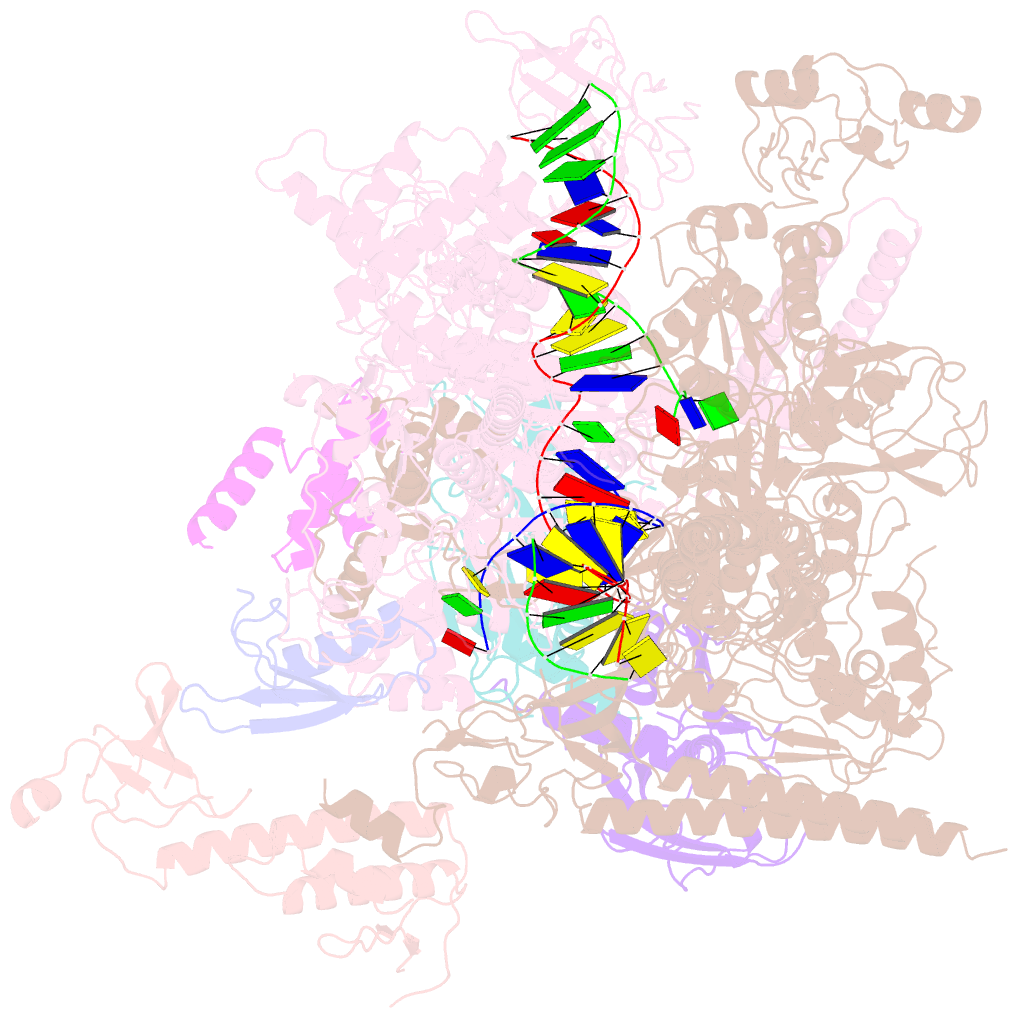
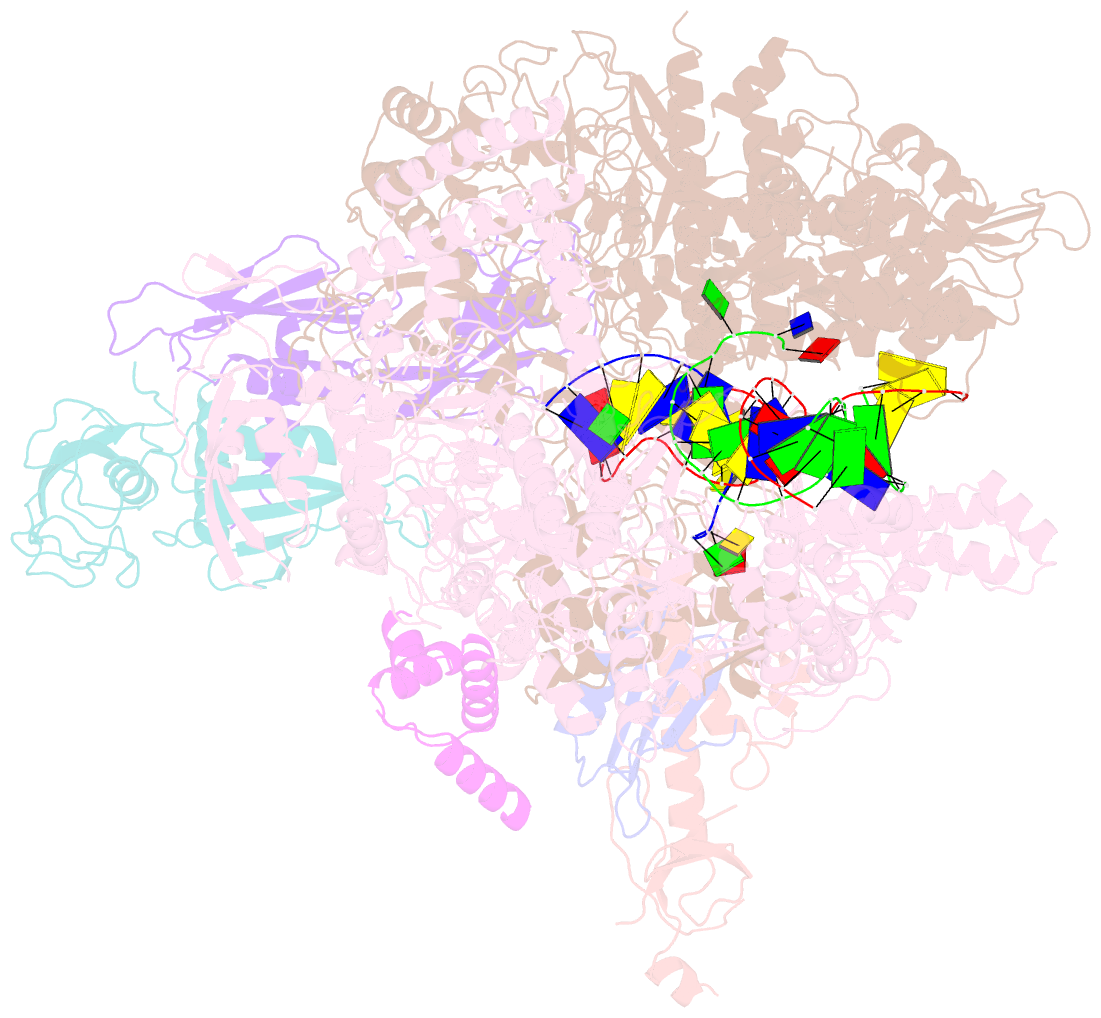
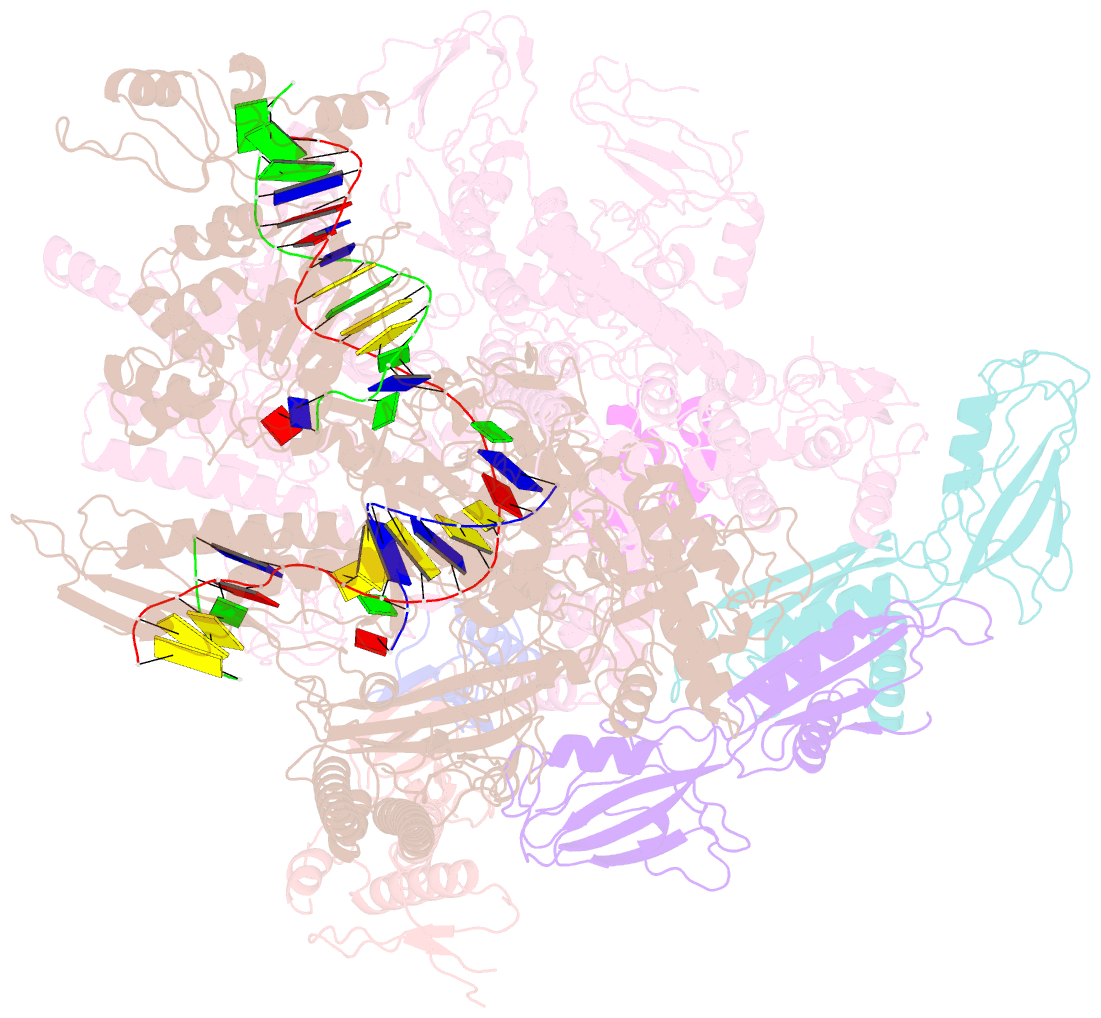
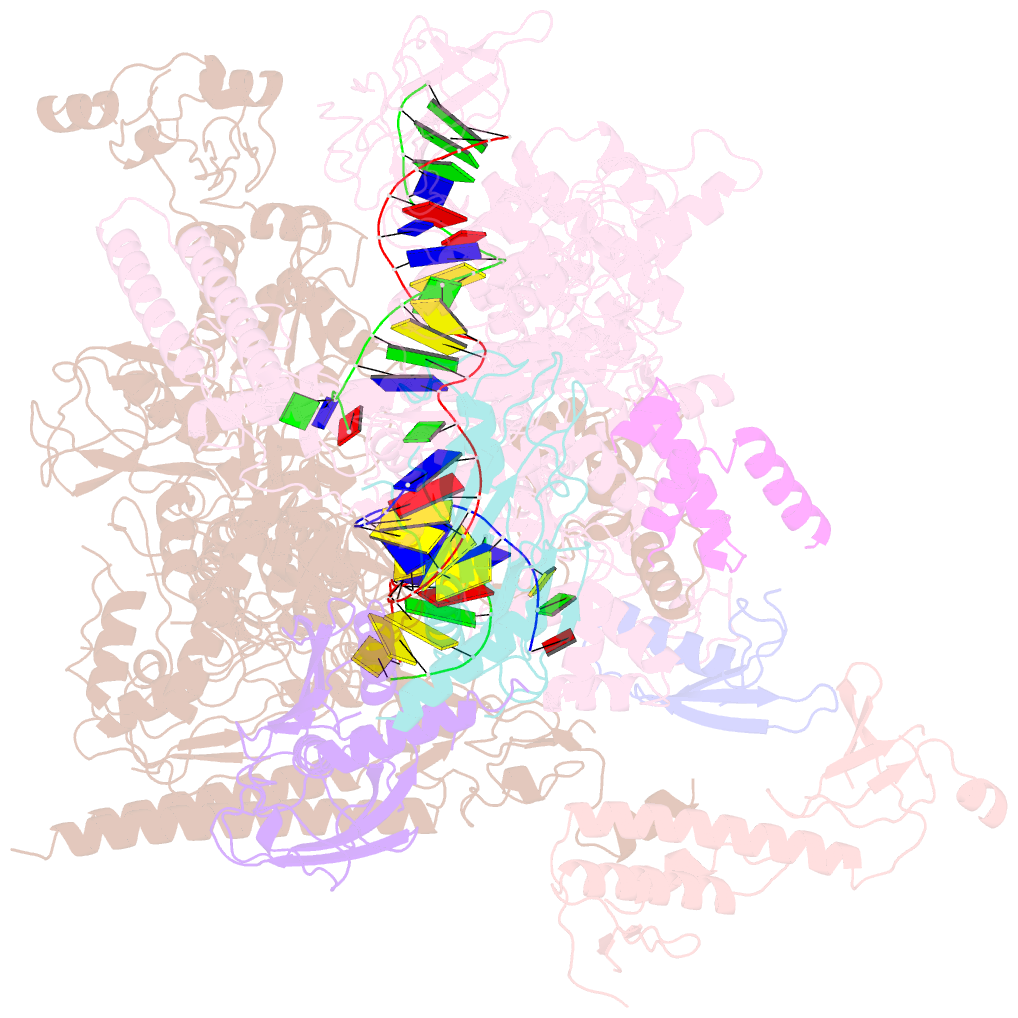
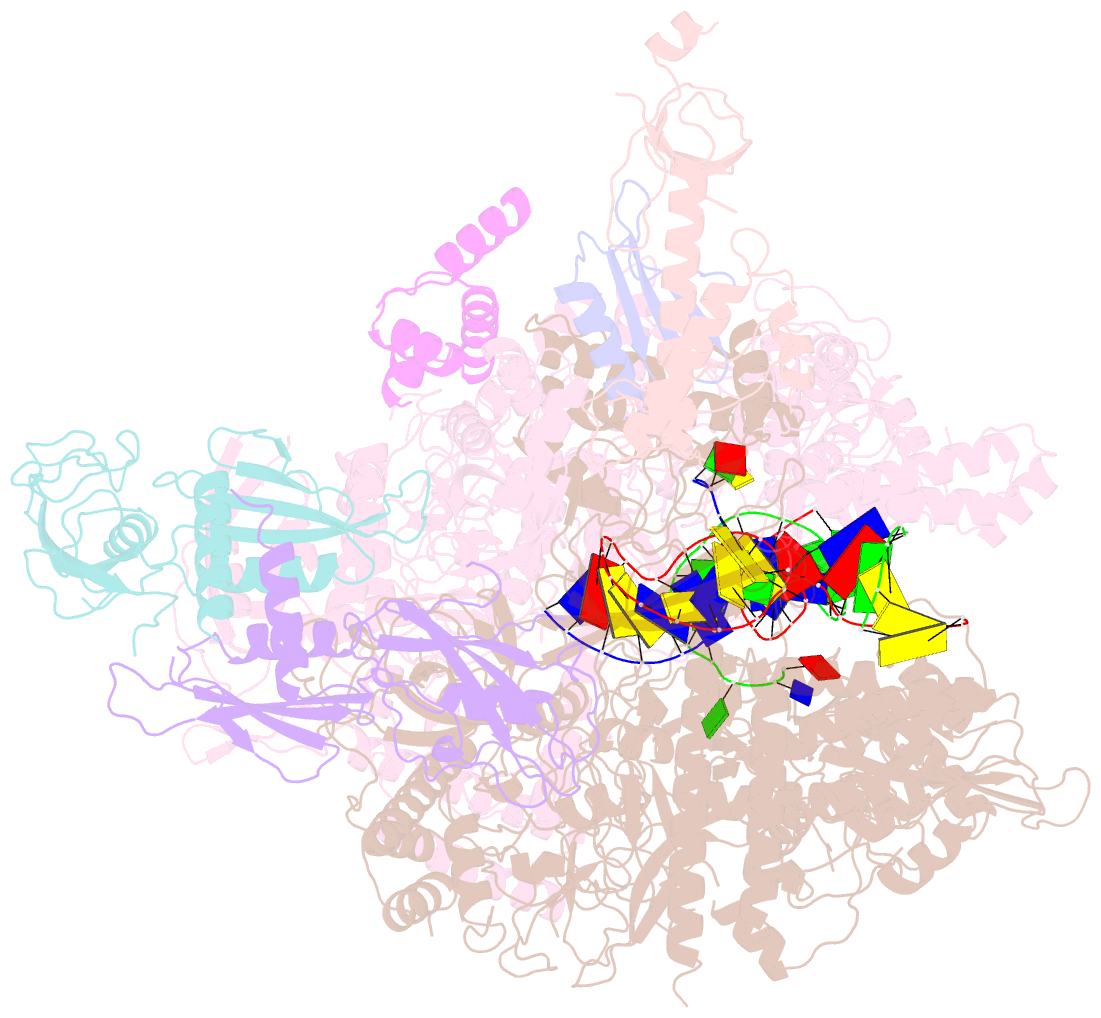
- cryo-EM structure of xanthomonos oryzae transcription elongation complex with nusa and the bacteriophage protein p7
- Reference
- You L, Shi J, Shen L, Li L, Fang C, Yu C, Cheng W, Feng Y, Zhang Y (2019): "Structural basis for transcription antitermination at bacterial intrinsic terminator." Nat Commun, 10, 3048. doi: 10.1038/s41467-019-10955-x.
- Abstract
- Bacteriophages typically hijack the host bacterial transcriptional machinery to regulate their own gene expression and that of the host bacteria. The structural basis for bacteriophage protein-mediated transcription regulation-in particular transcription antitermination-is largely unknown. Here we report the 3.4 Å and 4.0 Å cryo-EM structures of two bacterial transcription elongation complexes (P7-NusA-TEC and P7-TEC) comprising the bacteriophage protein P7, a master host-transcription regulator encoded by bacteriophage Xp10 of the rice pathogen Xanthomonas oryzae pv. Oryzae (Xoo) and discuss the mechanisms by which P7 modulates the host bacterial RNAP. The structures together with biochemical evidence demonstrate that P7 prevents transcription termination by plugging up the RNAP RNA-exit channel and impeding RNA-hairpin formation at the intrinsic terminator. Moreover, P7 inhibits transcription initiation by restraining RNAP-clamp motions. Our study reveals the structural basis for transcription antitermination by phage proteins and provides insights into bacterial transcription regulation.