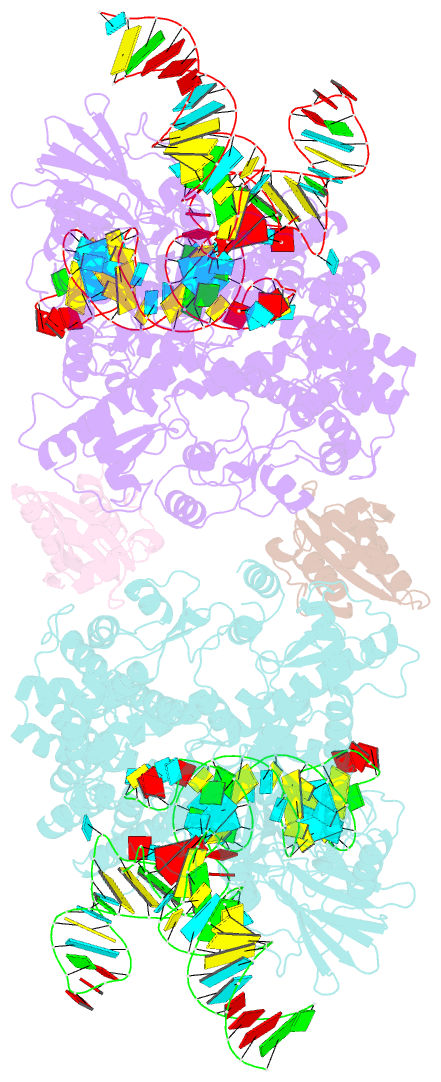
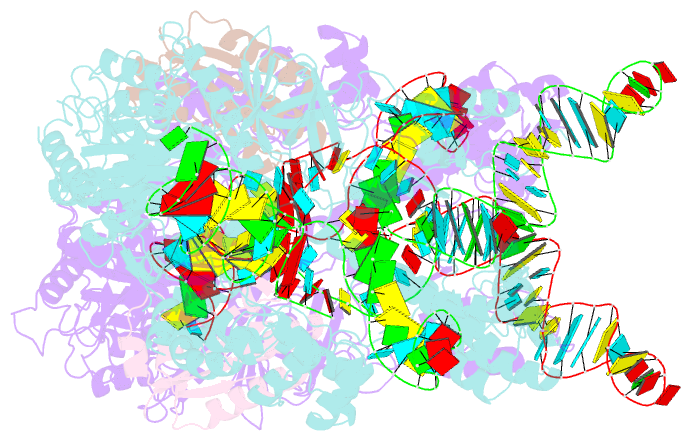
Summary information and primary citation
- PDB-id
- 6je9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-hydrolase inhibitor-RNA
- Method
- X-ray (3.46 Å)
- Summary
- Crystal structure of nme1cas9-sgrna dimer mediated by double protein inhibitor acriic3 monomers
- Reference
- Sun W, Yang J, Cheng Z, Amrani N, Liu C, Wang K, Ibraheim R, Edraki A, Huang X, Wang M, Wang J, Liu L, Sheng G, Yang Y, Lou J, Sontheimer EJ, Wang Y (2019): "Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States." Mol.Cell, 76, 938. doi: 10.1016/j.molcel.2019.09.025.
- Abstract
- High-resolution Cas9 structures have yet to reveal catalytic conformations due to HNH nuclease domain positioning away from the cleavage site. Nme1Cas9 and Nme2Cas9 are compact nucleases for in vivo genome editing. Here, we report structures of meningococcal Cas9 homologs in complex with sgRNA, dsDNA, or the AcrIIC3 anti-CRISPR protein. DNA-bound structures represent an early step of target recognition, a later HNH pre-catalytic state, the HNH catalytic state, and a cleaved-target-DNA-bound state. In the HNH catalytic state of Nme1Cas9, the active site is seen poised at the scissile phosphodiester linkage of the target strand, providing a high-resolution view of the active conformation. The HNH active conformation activates the RuvC domain. Our structures explain how Nme1Cas9 and Nme2Cas9 read distinct PAM sequences and how AcrIIC3 inhibits Nme1Cas9 activity. These structures provide insights into Cas9 domain rearrangements, guide-target engagement, cleavage mechanism, and anti-CRISPR inhibition, facilitating the optimization of these genome-editing platforms.