Summary information and primary citation
- PDB-id
- 6jnx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription,DNA,RNA
- Method
- cryo-EM (4.08 Å)
- Summary
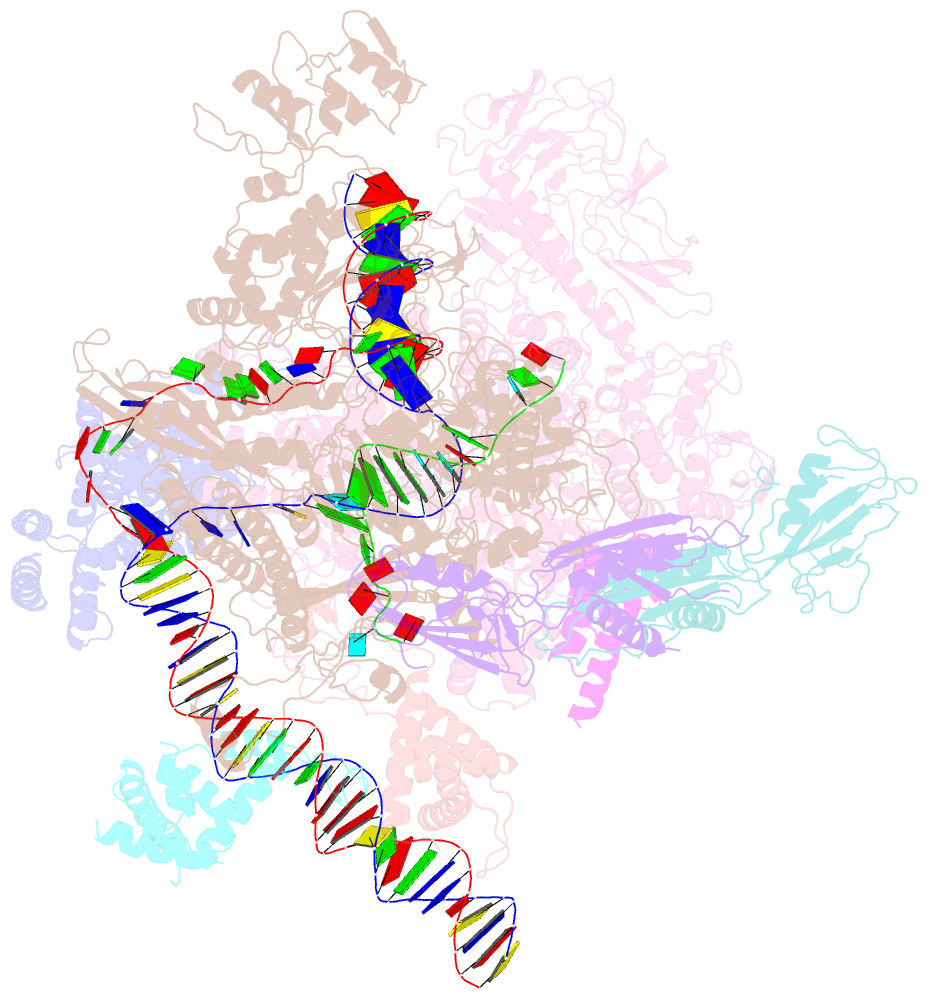
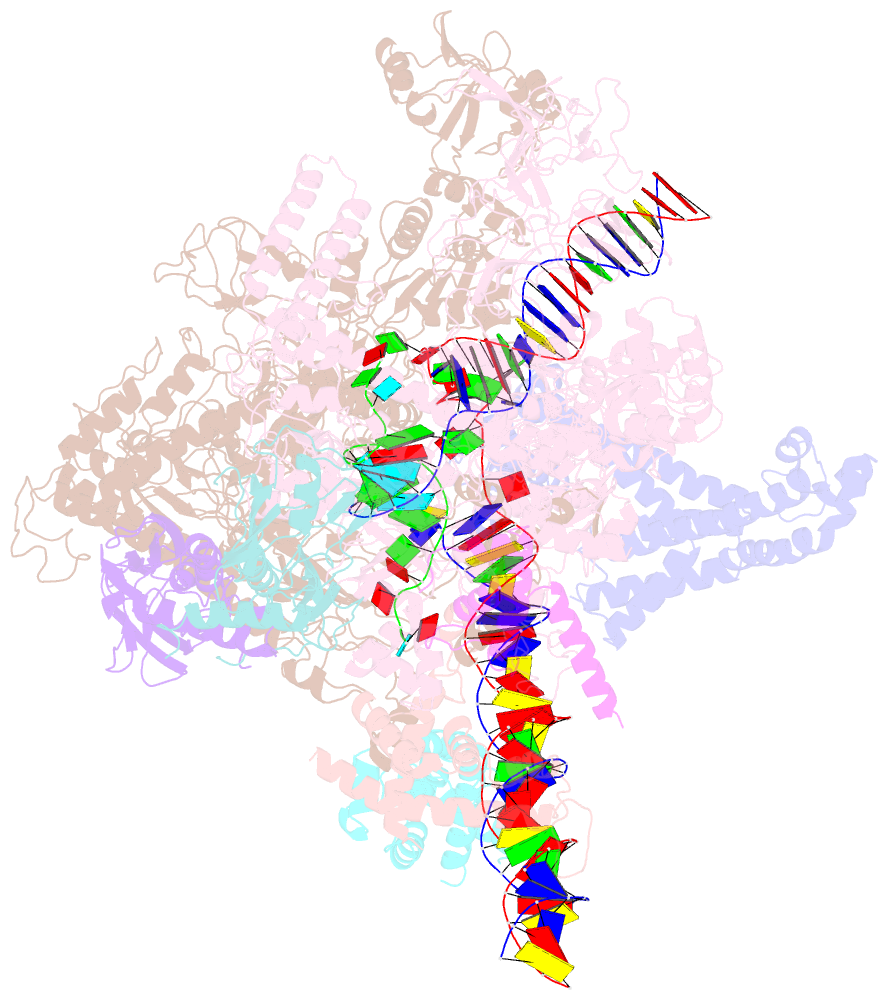
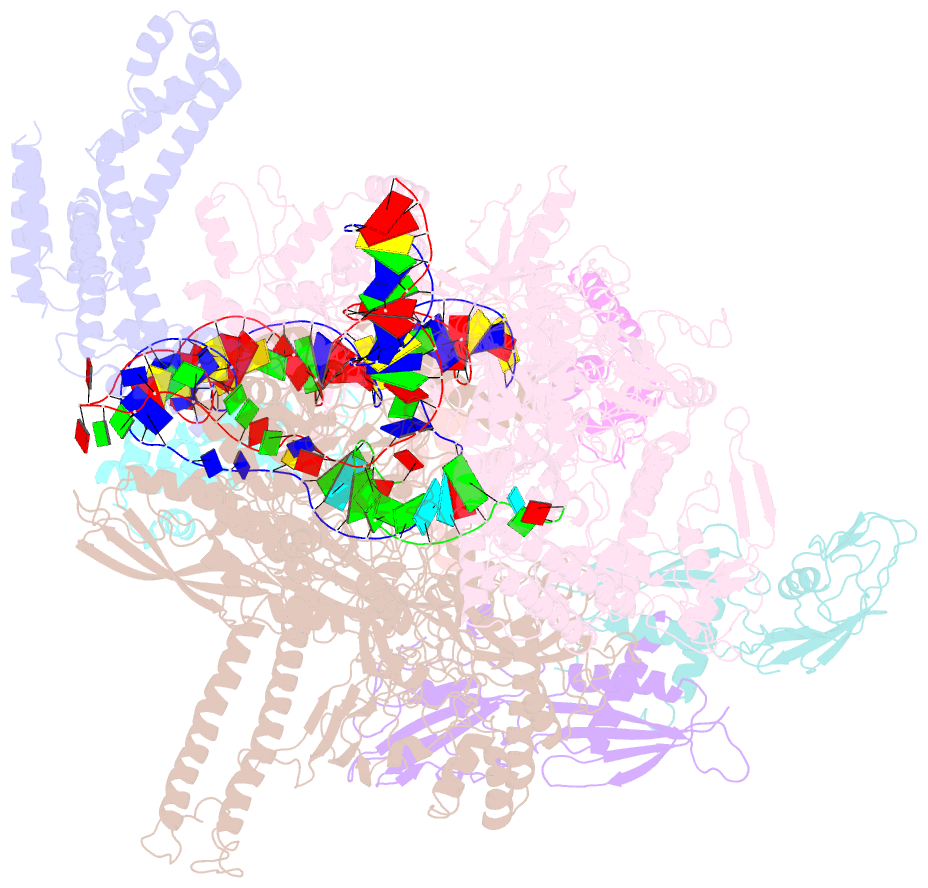
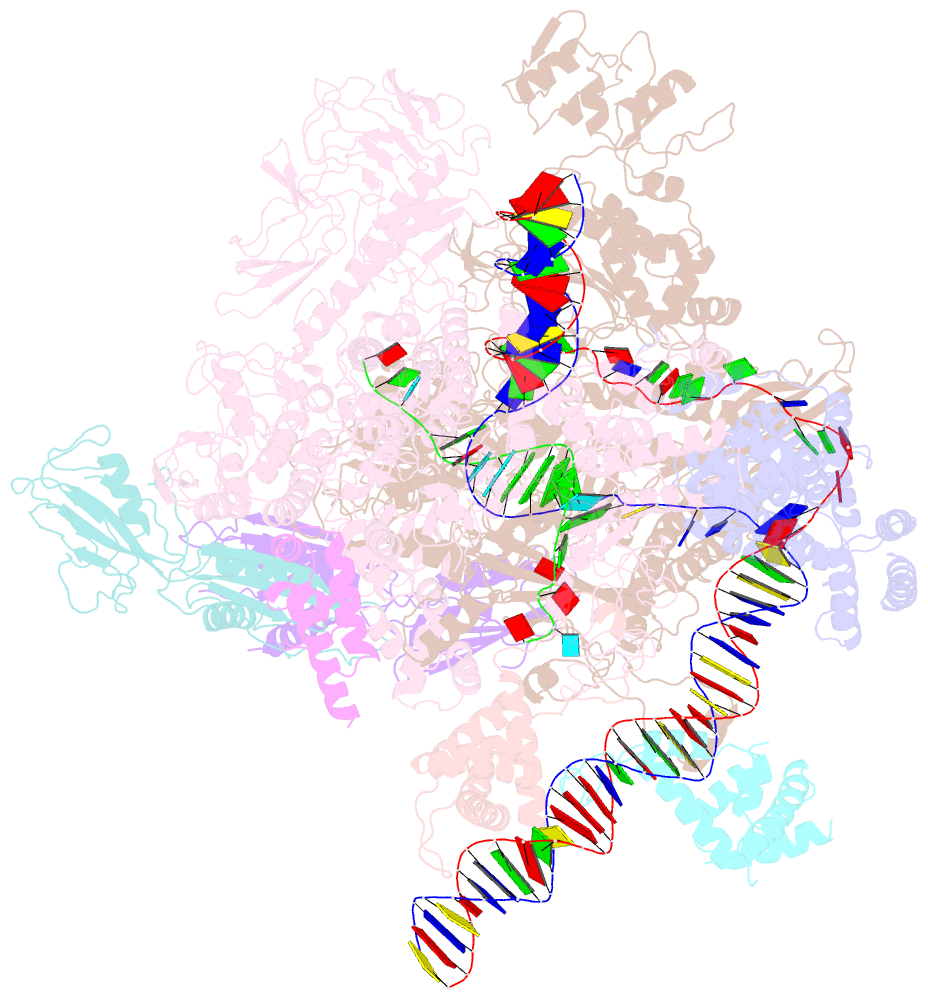
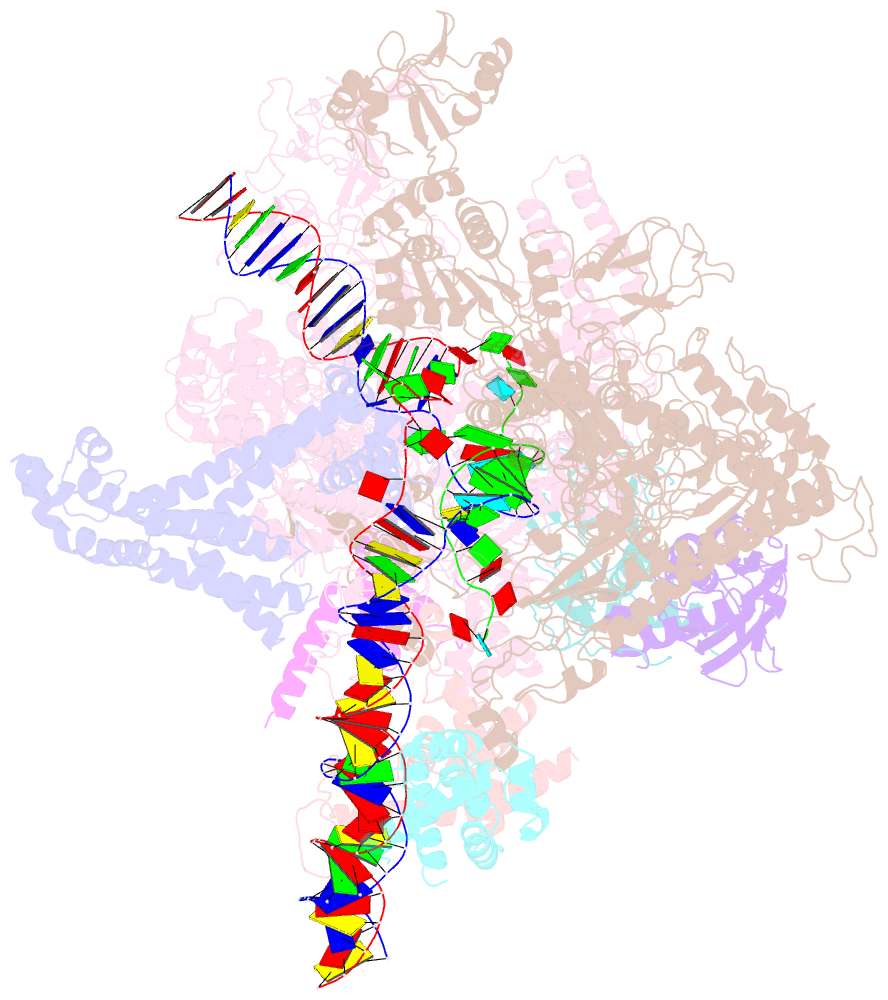
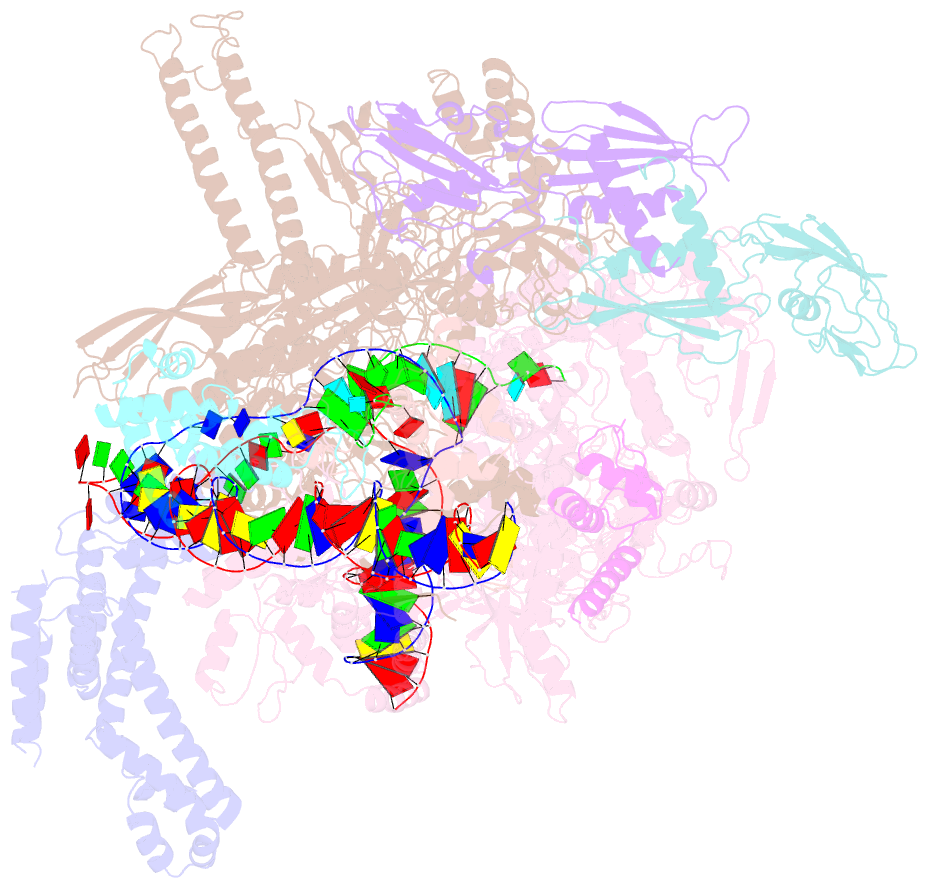
- cryo-EM structure of a q-engaged arrested complex
- Reference
- Shi J, Gao X, Tian T, Yu Z, Gao B, Wen A, You L, Chang S, Zhang X, Zhang Y, Feng Y (2019): "Structural basis of Q-dependent transcription antitermination." Nat Commun, 10, 2925. doi: 10.1038/s41467-019-10958-8.
- Abstract
- Bacteriophage Q protein engages σ-dependent paused RNA polymerase (RNAP) by binding to a DNA site embedded in late gene promoter and renders RNAP resistant to termination signals. Here, we report a single-particle cryo-electron microscopy (cryo-EM) structure of an intact Q-engaged arrested complex. The structure reveals key interactions responsible for σ-dependent pause, Q engagement, and Q-mediated transcription antitermination. The structure shows that two Q protomers (QI and QII) bind to a direct-repeat DNA site and contact distinct elements of the RNA exit channel. Notably, QI forms a narrow ring inside the RNA exit channel and renders RNAP resistant to termination signals by prohibiting RNA hairpin formation in the RNA exit channel. Because the RNA exit channel is conserved among all multisubunit RNAPs, it is likely to serve as an important contact site for regulators that modify the elongation properties of RNAP in other organisms, as well.