Summary information and primary citation
- PDB-id
- 6k2j; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.4 Å)
- Summary
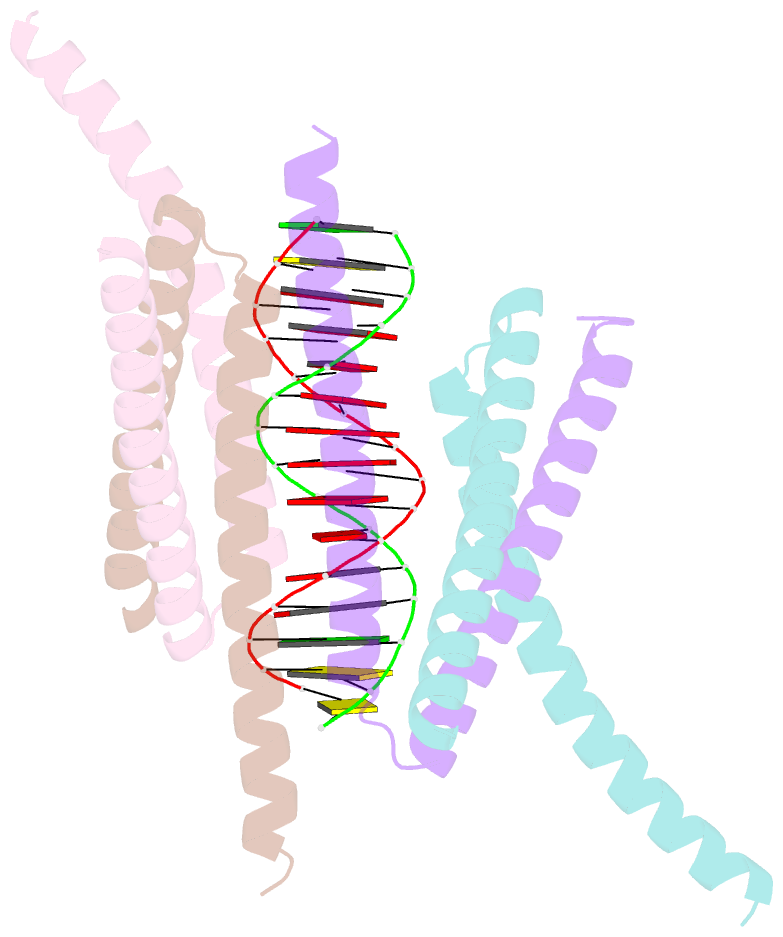
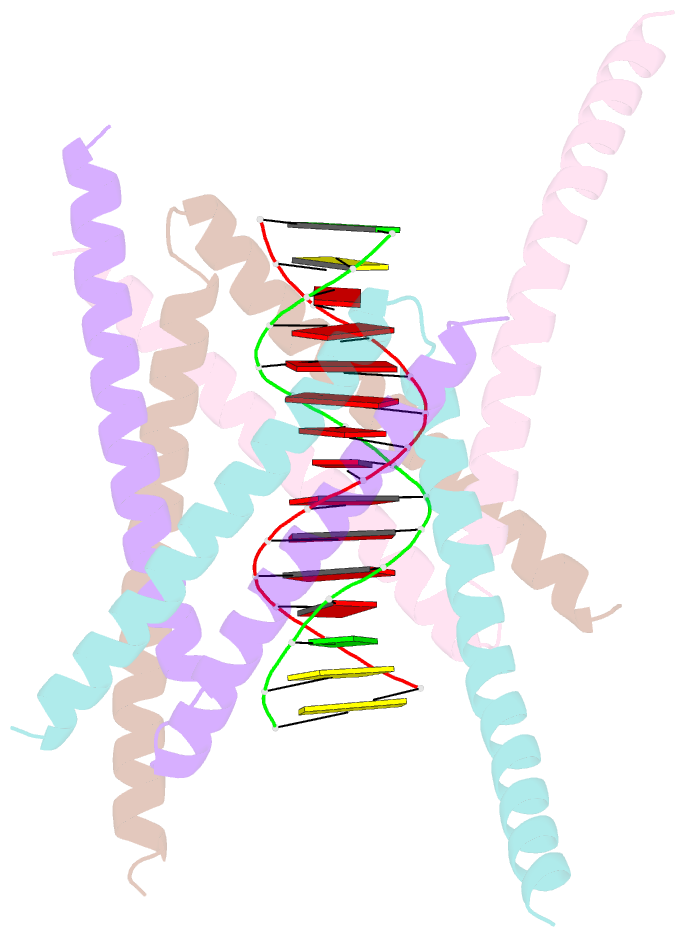
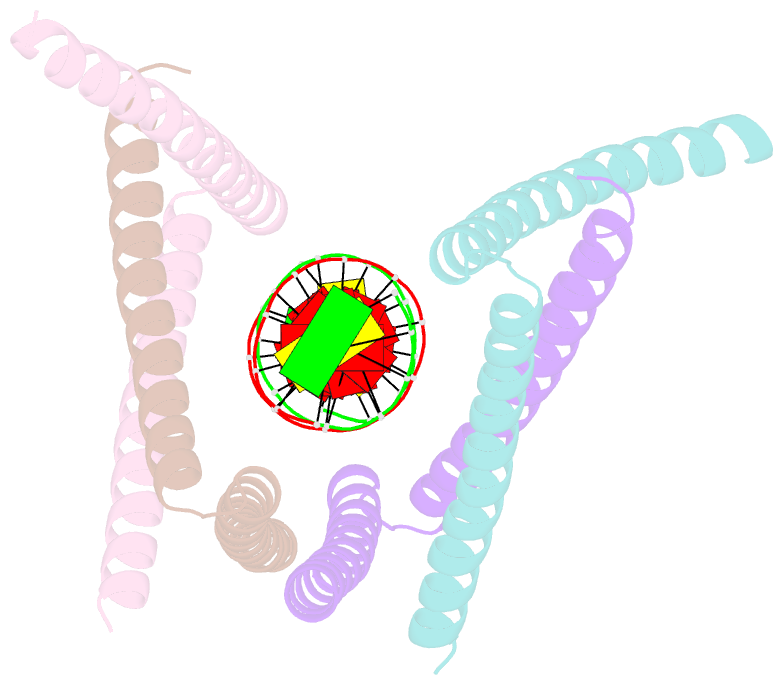
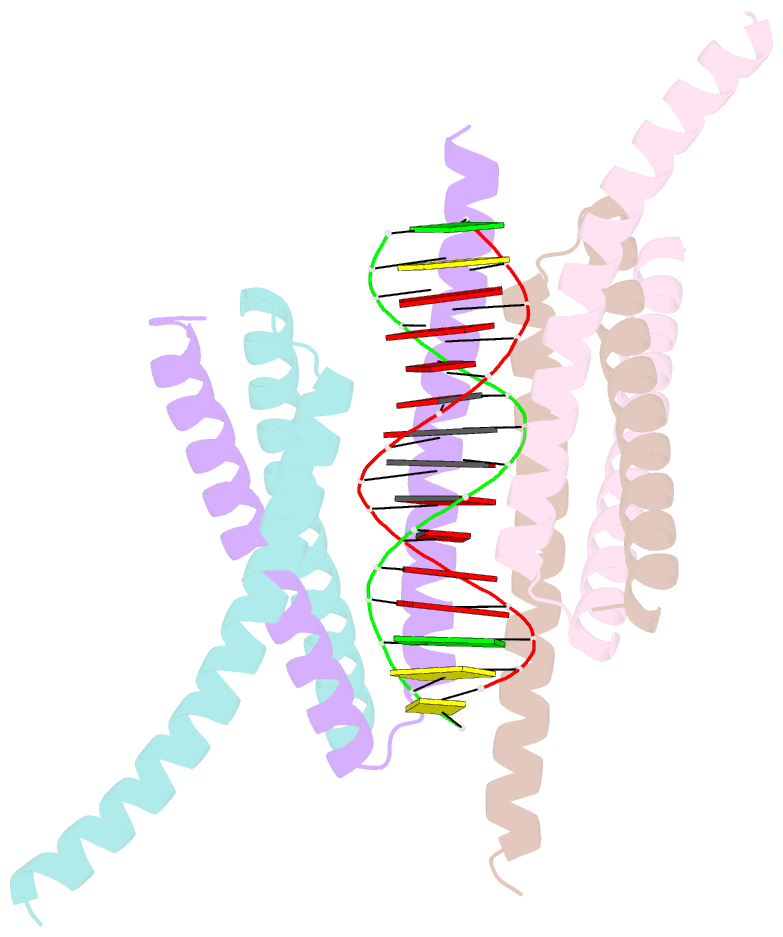
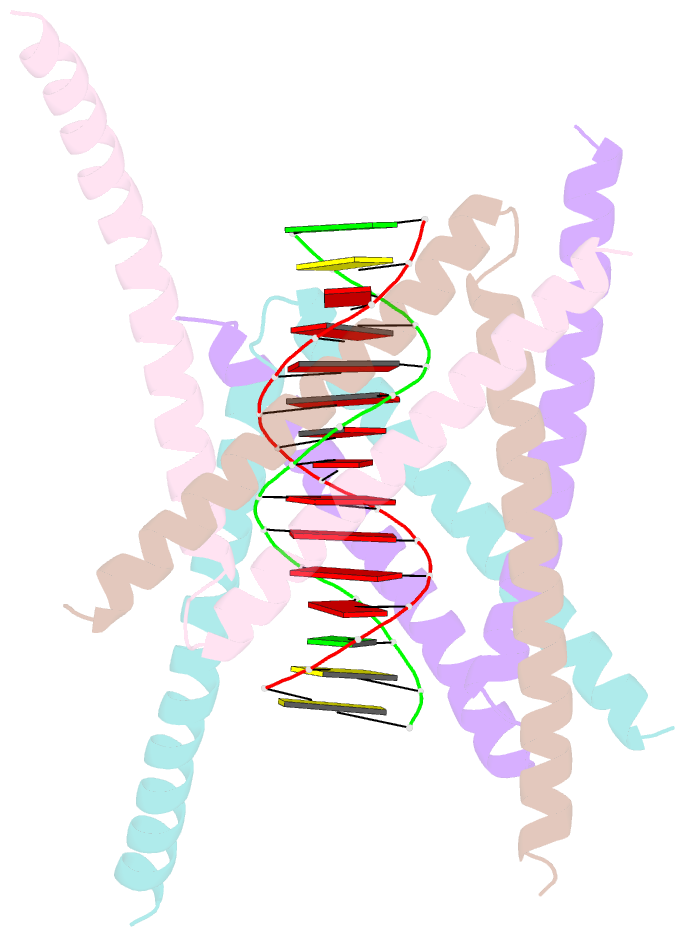
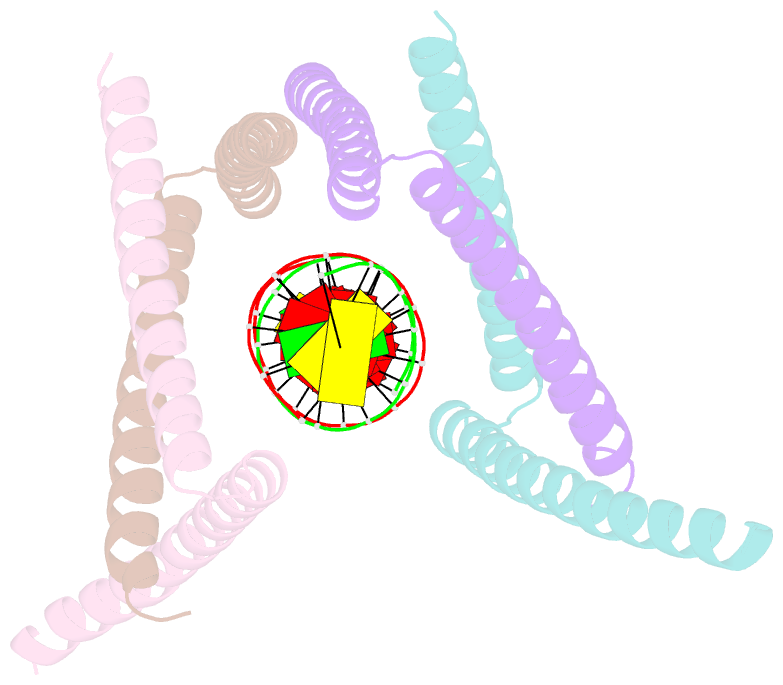
- Crystal structure of the DNA complex of c. crescentus gapr
- Reference
- Huang Q, Duan B, Dong X, Fan S, Xia B (2020): "GapR binds DNA through dynamic opening of its tetrameric interface." Nucleic Acids Res., 48, 9372-9386. doi: 10.1093/nar/gkaa644.
- Abstract
- GapR is a nucleoid-associated protein that is an essential regulator of chromosome replication in the cell cycle model Caulobacter crescentus. Here, we demonstrate that free GapR is a homotetramer, but not a dimer as previously reported (Guo et al., Cell 175: 583-597, 2018). We have determined the crystal structure of GapR in complex with a 10-bp A-tract DNA, which has an open tetrameric conformation, different from the closed clamp conformation in the previously reported crystal structure of GapR/DNA complex. The free GapR adopts multiple conformations in dynamic exchange equilibrium, with the major conformation resembling the closed tetrameric conformation, while the open tetrameric conformation is a representative of minor conformers. As it is impossible for the circular genomic DNA to get into the central DNA binding tunnel of the major conformation, we propose that GapR initially binds DNA through the open conformation, and then undergoes structural rearrangement to form the closed conformation which fully encircles the DNA. GapR prefers to bind DNA with 10-bp consecutive A/T base pairs nonselectively (Kd ∼12 nM), while it can also bind GC-rich DNA sequence with a reasonable affinity of about 120 nM. Besides, our results suggest that GapR binding results in widening the minor groove of AT-rich DNA, instead of overtwisting DNA.