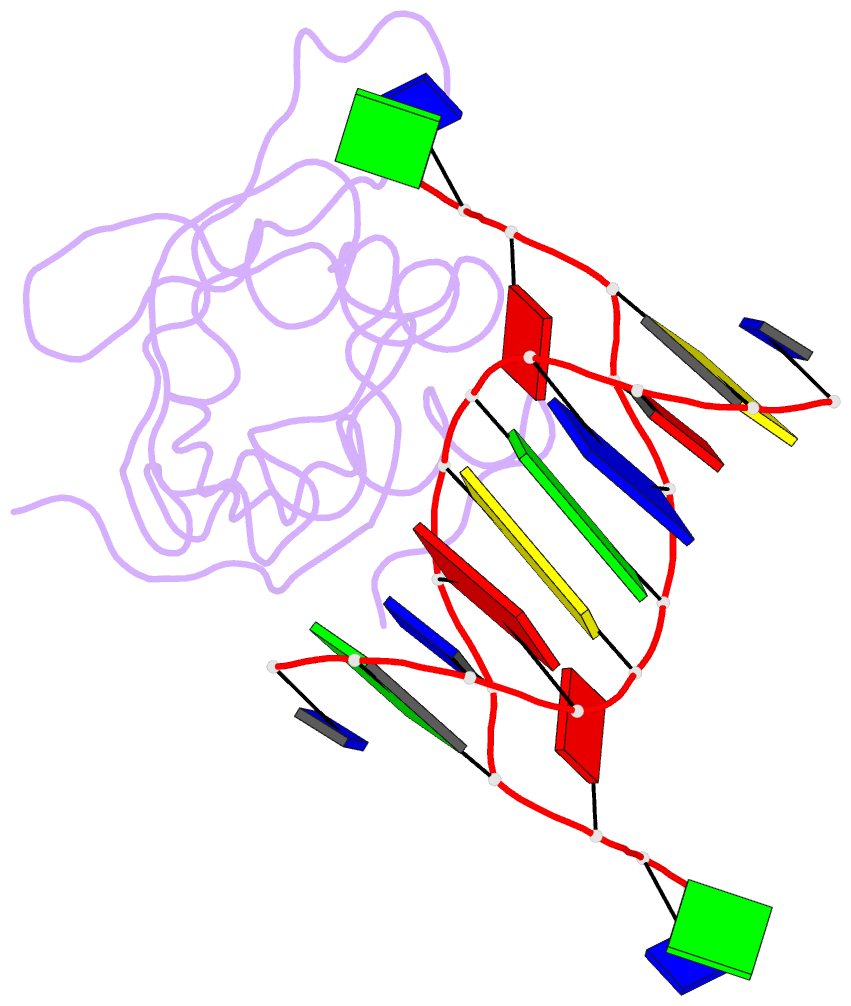
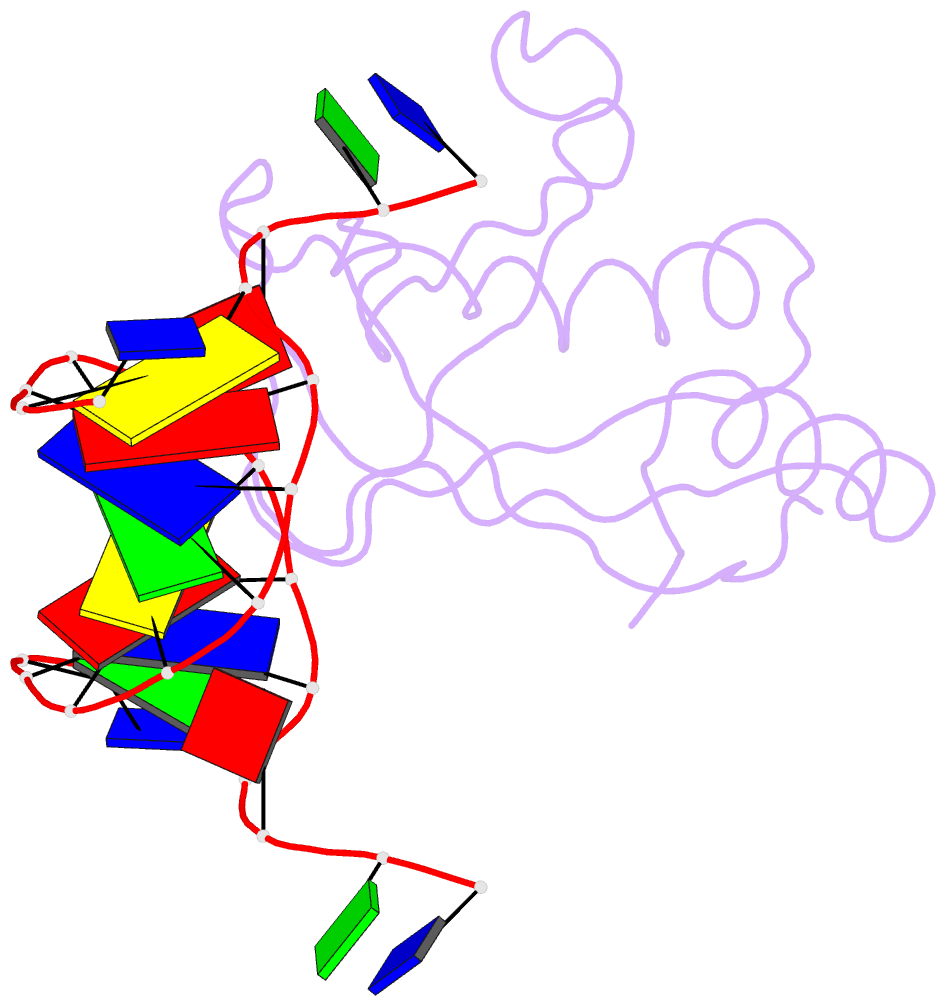
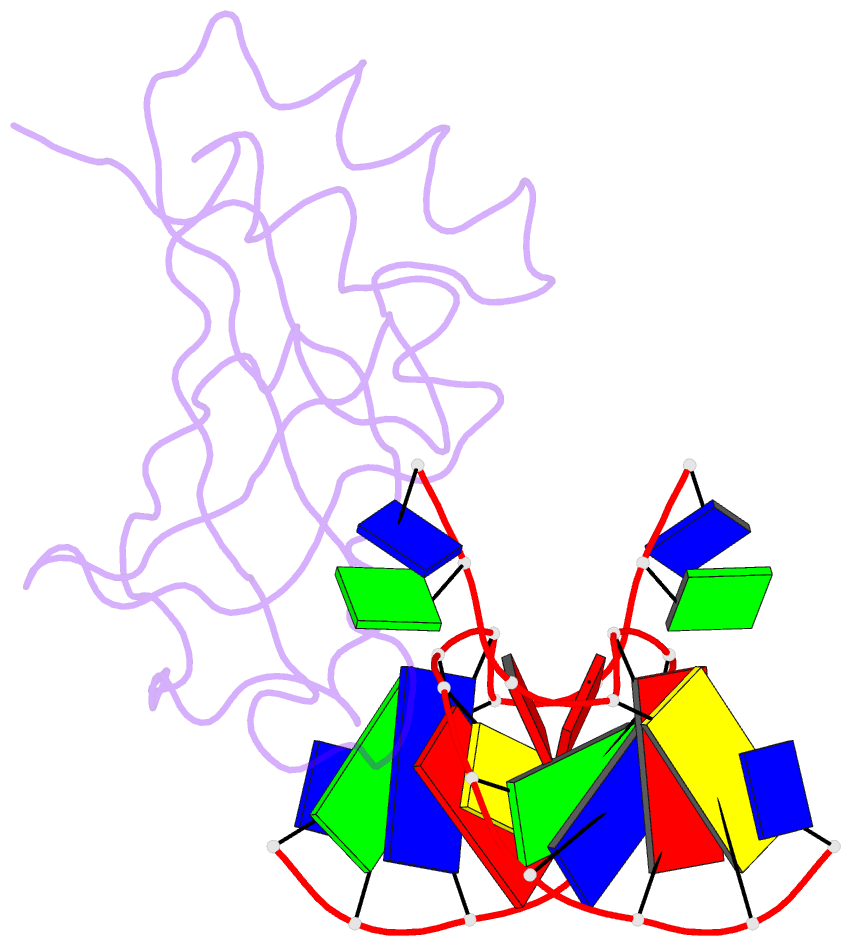
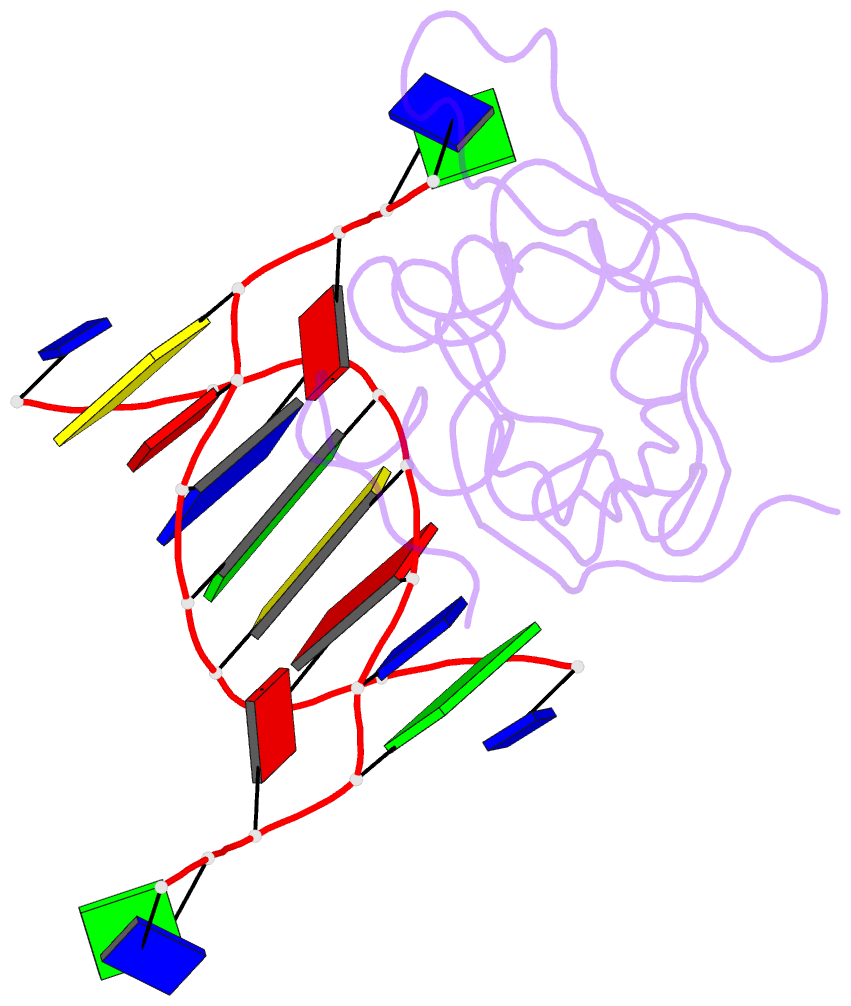
Summary information and primary citation
- PDB-id
- 6kcs; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.1 Å)
- Summary
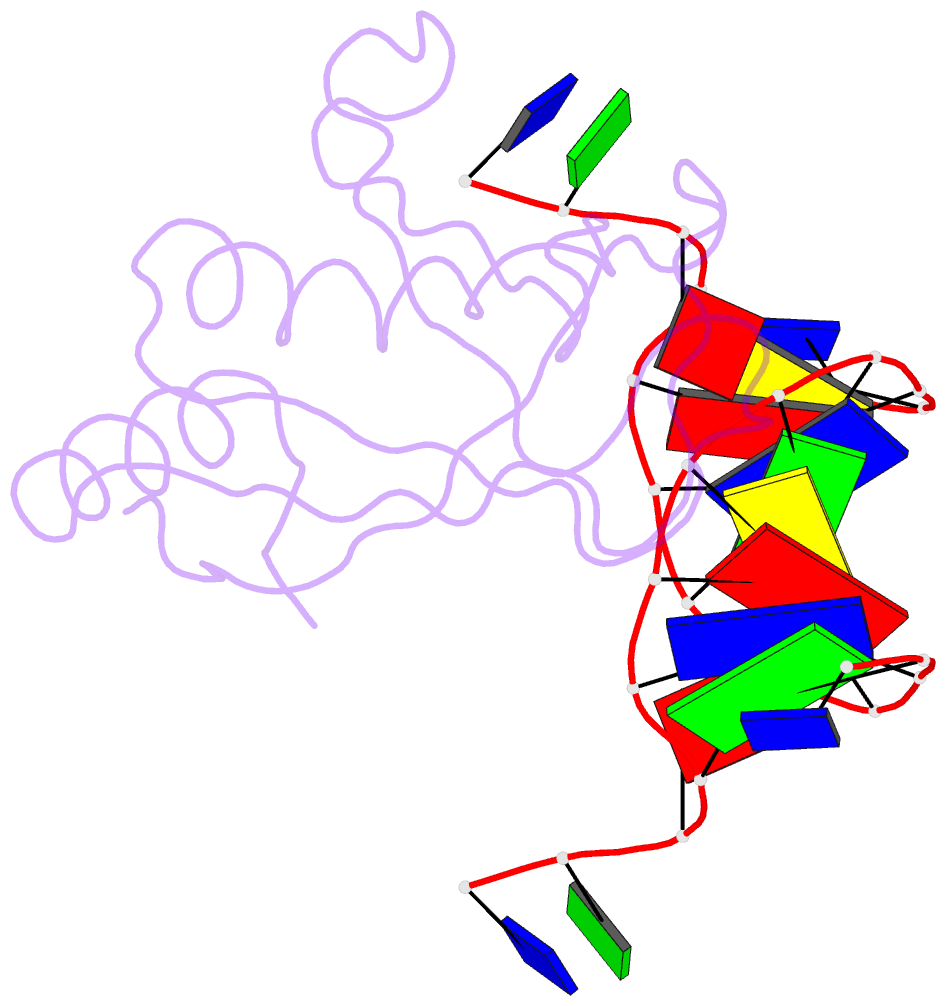
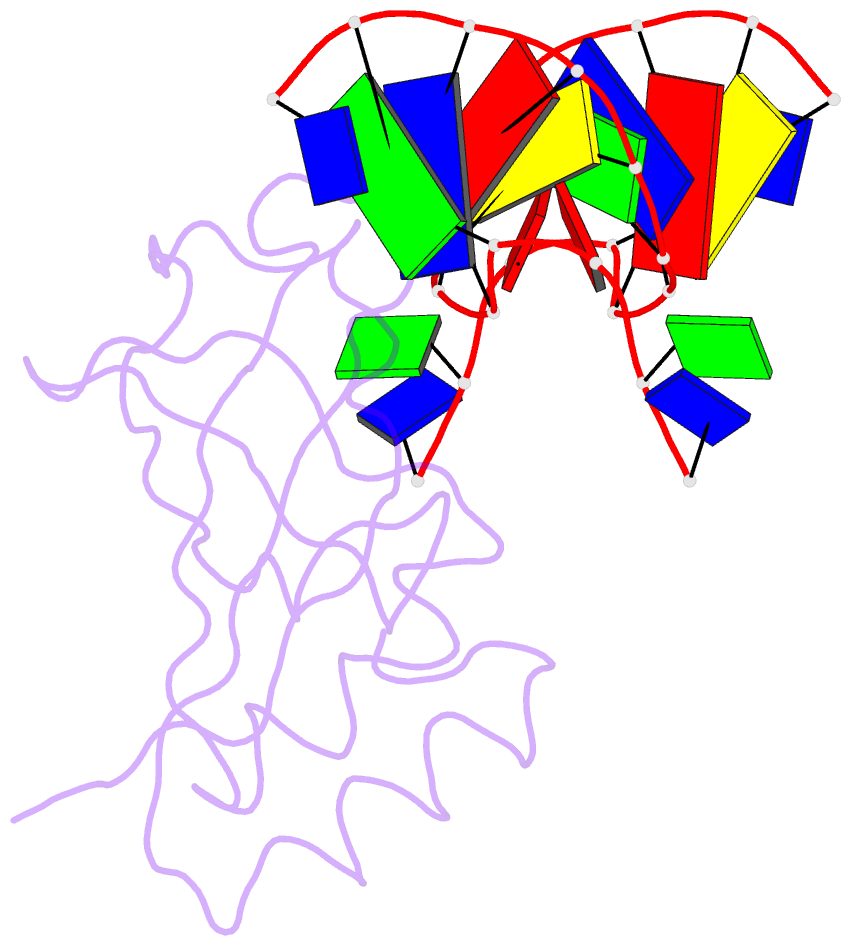
- Crystal structure of hiran domain of hltf in complex with duplex DNA
- Reference
- Hishiki A, Sato M, Hashimoto H (2020): "Structure of HIRAN domain of human HLTF bound to duplex DNA provides structural basis for DNA unwinding to initiate replication fork regression." J.Biochem., 167, 597-602. doi: 10.1093/jb/mvaa008.
- Abstract
- Replication fork regression is a mechanism to rescue a stalled fork by various replication stresses, such as DNA lesions. Helicase-like transcription factor, a SNF2 translocase, plays a central role in the fork regression and its N-terminal domain, HIRAN (HIP116 and Rad5 N-terminal), binds the 3'-hydroxy group of single-stranded DNA. Furthermore, HIRAN is supposed to bind double-stranded DNA (dsDNA) and involved in strand separation in the fork regression, whereas structural basis for mechanisms underlying dsDNA binding and strand separation by HIRAN are still unclear. Here, we report the crystal structure of HIRAN bound to duplex DNA. The structure reveals that HIRAN binds the 3'-hydroxy group of DNA and unexpectedly unwinds three nucleobases of the duplex. Phe-142 is involved in the dsDNA binding and the strand separation. In addition, the structure unravels the mechanism underlying sequence-independent recognition for purine bases by HIRAN, where the N-glycosidic bond adopts syn conformation. Our findings indicate direct involvement of HIRAN in the fork regression by separating of the daughter strand from the parental template.