Summary information and primary citation
- PDB-id
- 6kdi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system
- Method
- X-ray (2.7 Å)
- Summary
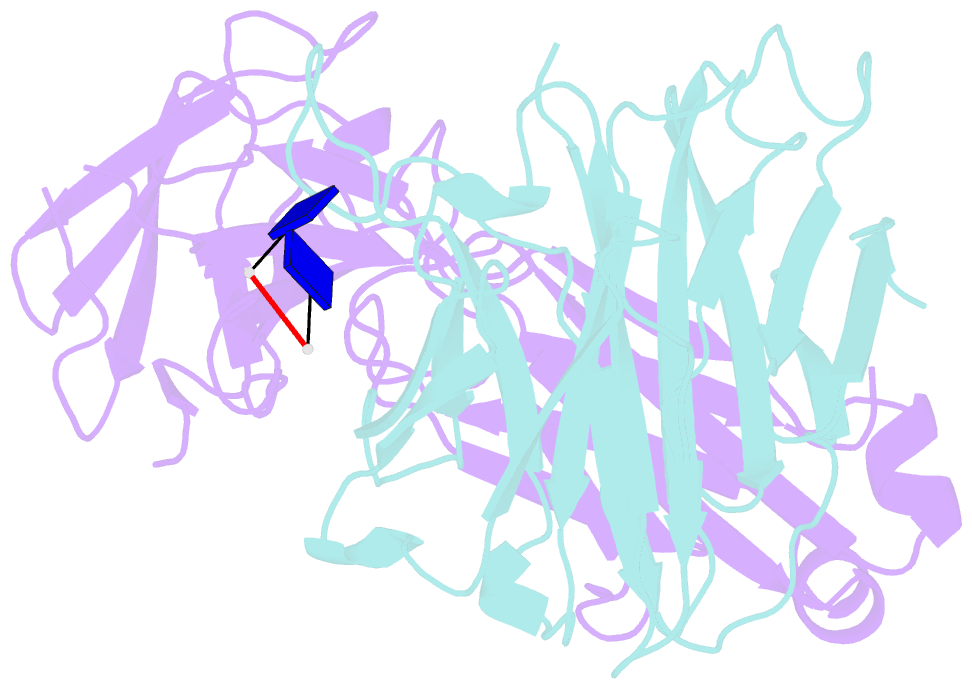
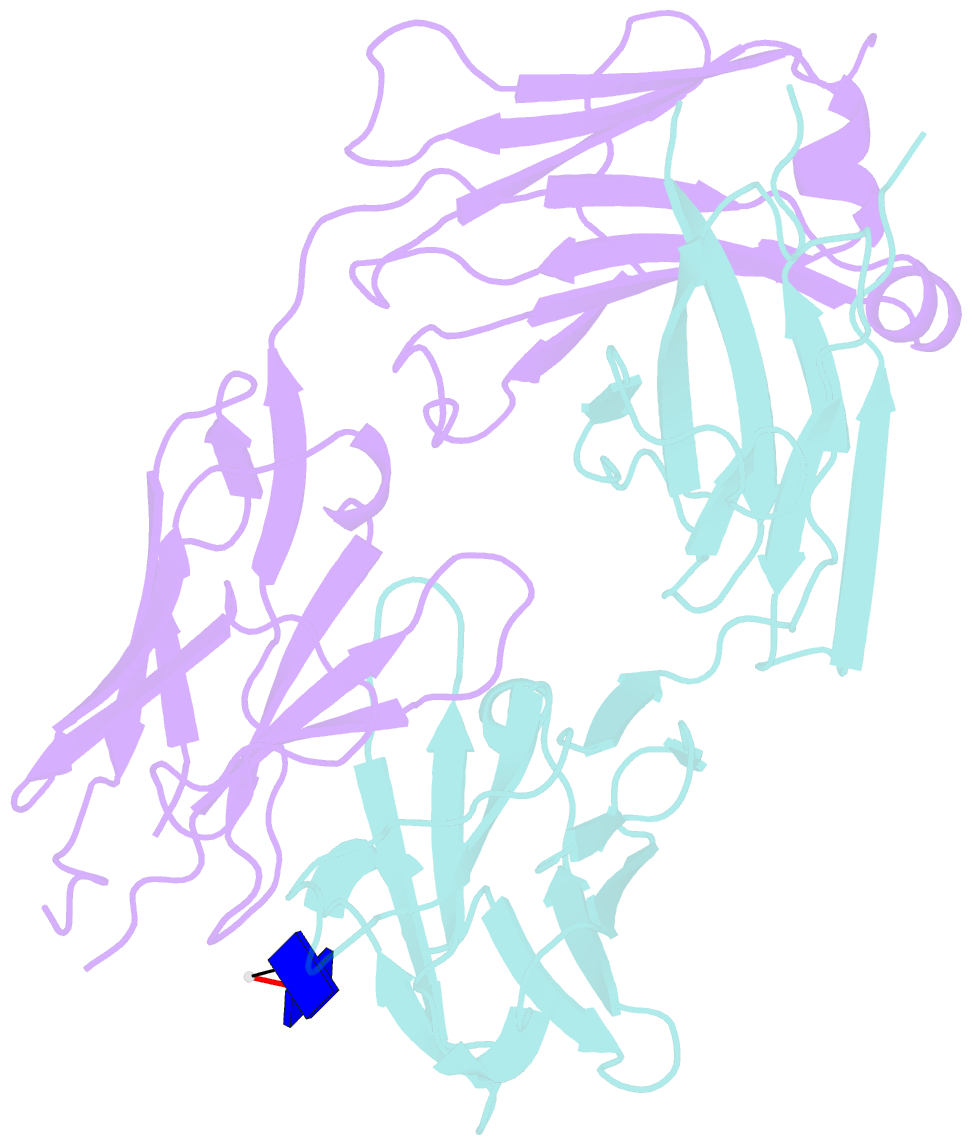
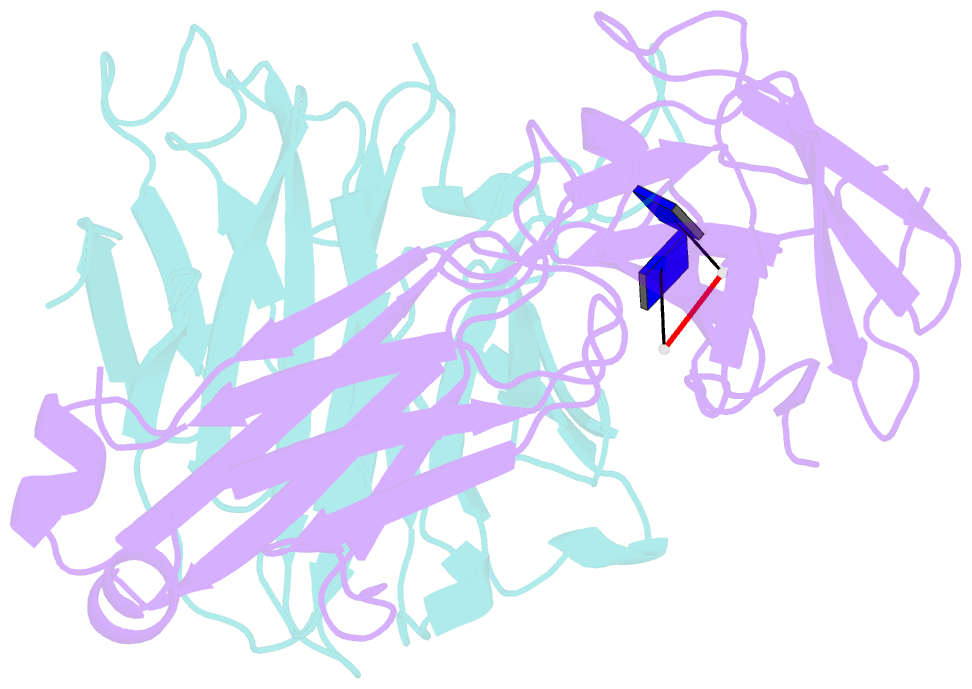
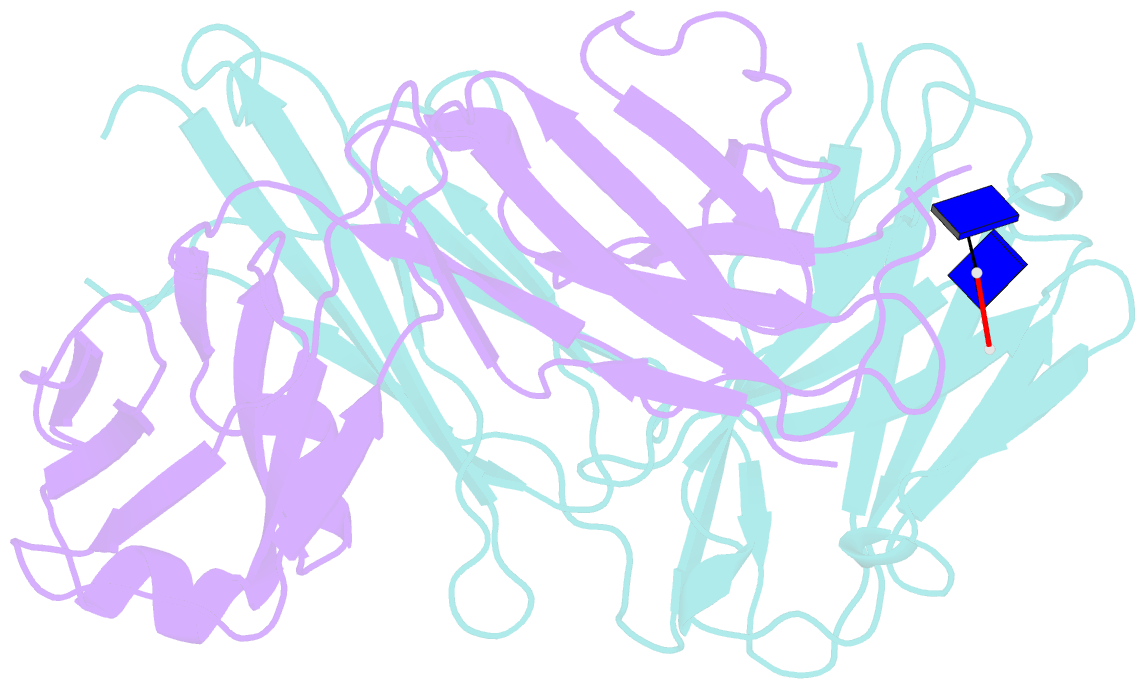
- Antibody 64m-5 fab including isoasp in complex with dt(6-4)t
- Reference
- Yokoyama H, Mizutani R, Noguchi S, Hayashida N (2019): "Structural and biochemical basis of the formation of isoaspartate in the complementarity-determining region of antibody 64M-5 Fab." Sci Rep, 9, 18494. doi: 10.1038/s41598-019-54918-0.
- Abstract
- The formation of the isoaspartate (isoAsp) is one of spontaneous degradation processes of proteins, affecting their stability and activity. Here, we report for the first time the crystal structures of an antibody Fab that contains isoAsp in the complementarity-determining region (CDR), along with biochemical studies to detect isoAsp. By comparing the elution profiles of cation-exchange chromatography, it was clarified that the antibody 64M-5 Fab is converted from the normal form to isoAsp form spontaneously and time-dependently under physiological conditions. The isoAsp residue was identified with tryptic peptide mapping, N-terminal sequencing, and the protein isoaspartyl methyltransferase assay. Based on the fluorescence quenching method, the isoAsp form of 64M-5 Fab shows a one order of magnitude lower binding constant for its dinucleotide ligand dT(6-4)T than the normal form. According to the structure of the isoAsp form, the conformation of CDR L1 is changed from the normal form to isoAsp form; the loss of hydrogen bonds involving the Asn28L side-chain, and structural conversion of the β-turn from type I to type II'. The formation of isoAsp leads to a large displacement of the side chain of His27dL, and decreased electrostatic interactions with the phosphate group of dT(6-4)T. Such structural changes should be responsible for the lower affinity of the isoAsp form for dT(6-4)T than the normal form. These findings may provide insight into neurodegenerative diseases (NDDs) and related diseases caused by misfolded proteins.