Summary information and primary citation
- PDB-id
- 6kdv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.11 Å)
- Summary
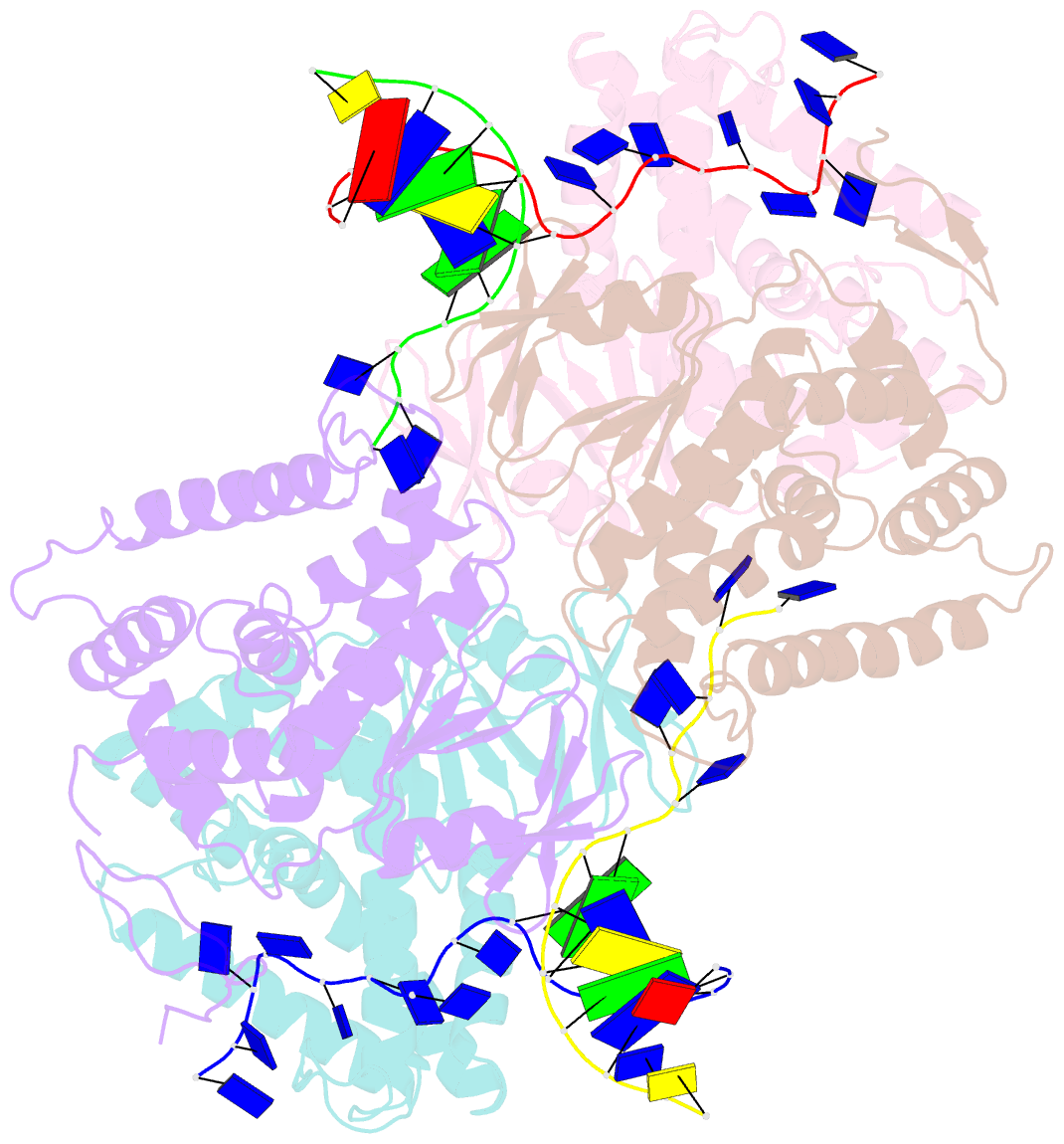
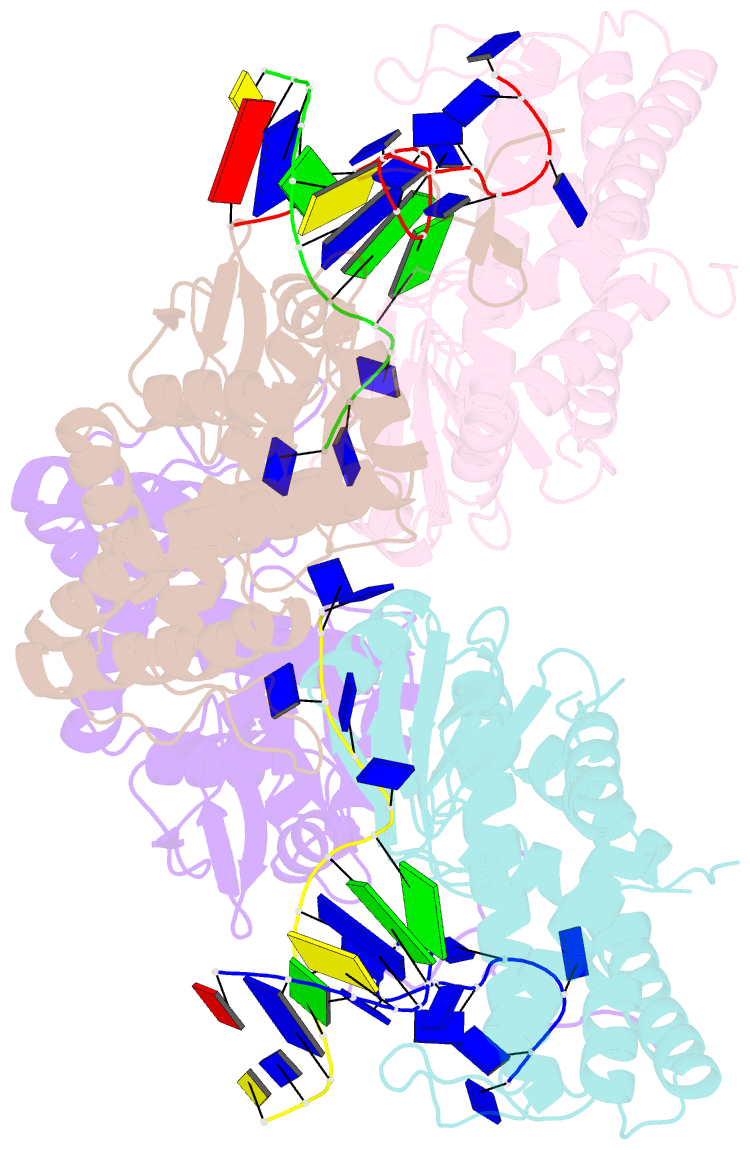
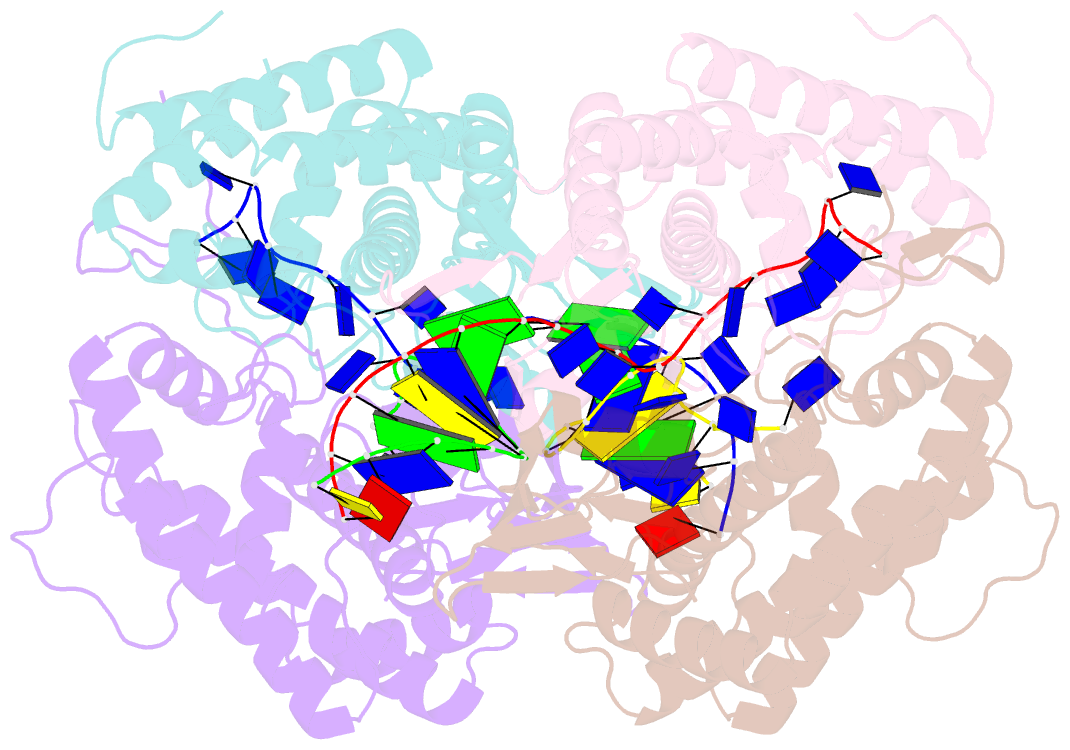
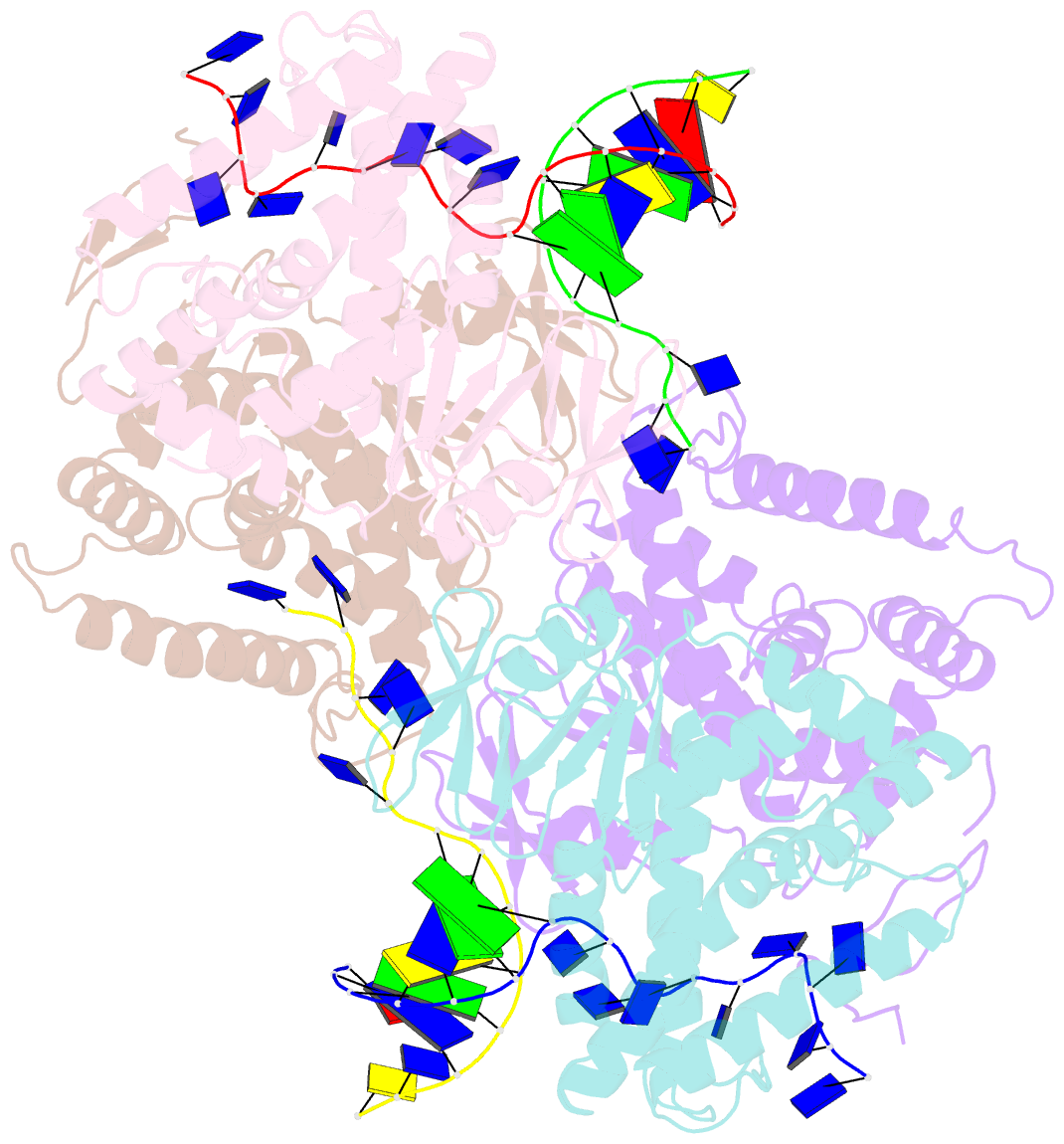
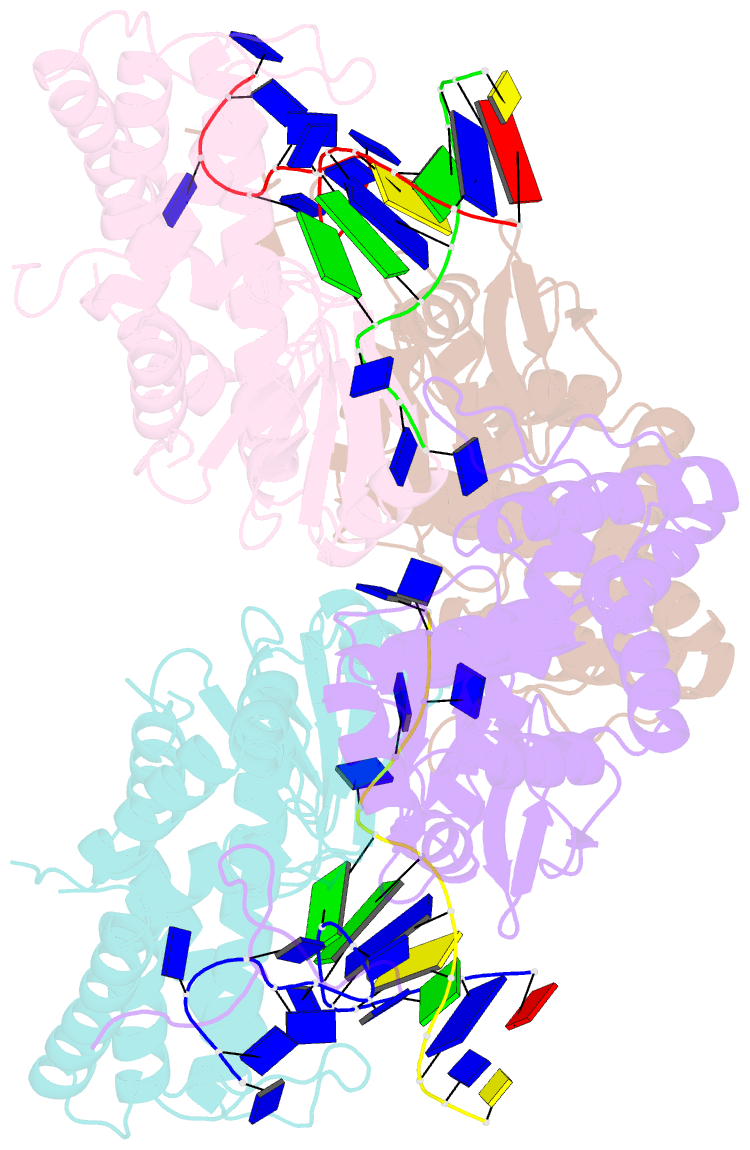
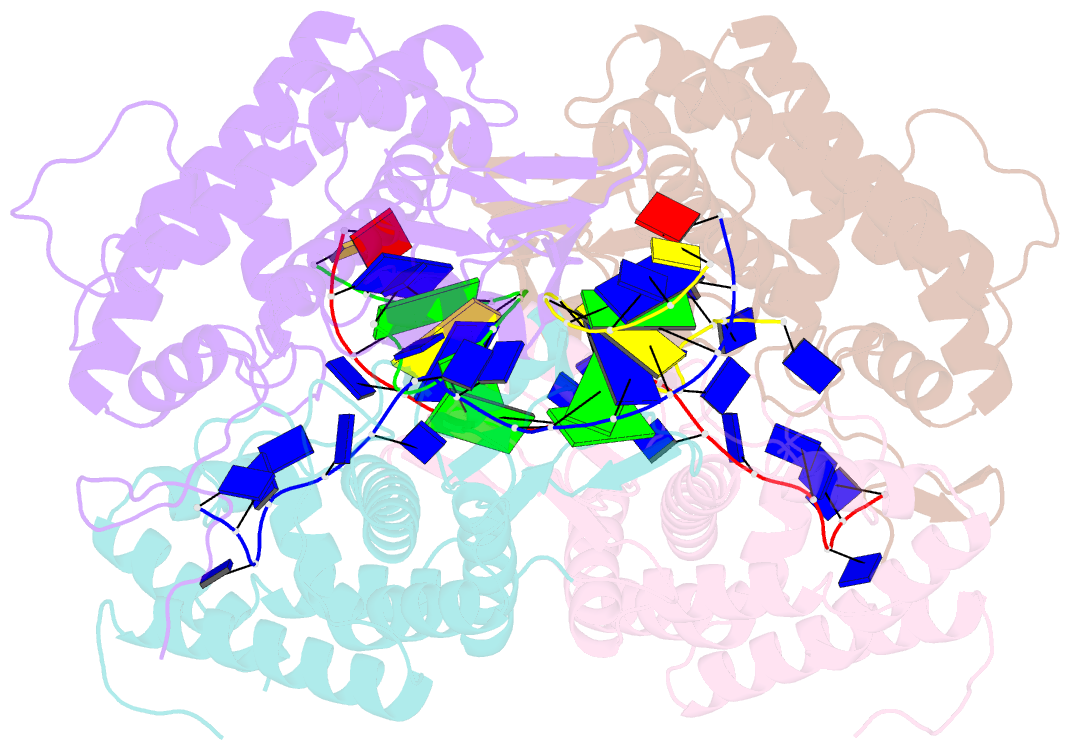
- Crystal structure of ttcas1-DNA complex
- Reference
- Yang J, Li J, Wang J, Sheng G, Wang M, Zhao H, Yang Y, Wang Y (2020): "Crystal structure of Cas1 in complex with branched DNA." Sci China Life Sci, 63, 516-528. doi: 10.1007/s11427-019-9827-x.
- Abstract
- Cas1 is a key component of the CRISPR adaptation complex, which captures and integrates foreign DNA into the CRISPR array, resulting in the generation of new spacers. We have determined crystal structures of Thermus thermophilus Cas1 involved in new spacer acquisition both in complex with branched DNA and in the free state. Cas1 forms an asymmetric dimer without DNA. Conversely, two asymmetrical dimers bound to two branched DNAs result in the formation of a DNA-mediated tetramer, dimer of structurally asymmetrical dimers, in which the two subunits markedly present different conformations. In the DNA binding complex, the N-terminal domain adopts different orientations with respect to the C-terminal domain in the two monomers that form the dimer. Substrate binding triggers a conformational change in the loop 164-177 segment. This loop is also involved in the 3' fork arm and 5' fork arm strand recognition in monomer A and B, respectively. This study provides important insights into the molecular mechanism of new spacer adaptation.