Summary information and primary citation
- PDB-id
- 6kks; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.15 Å)
- Summary
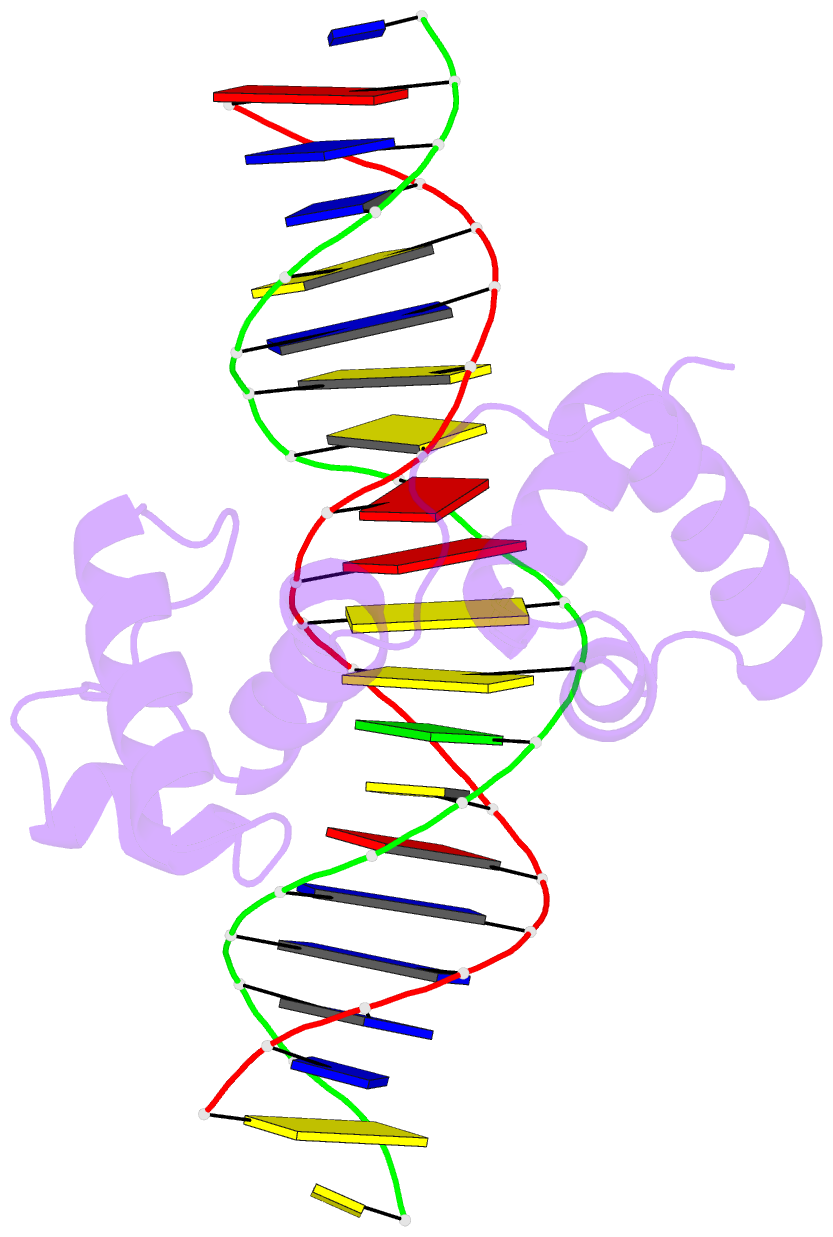
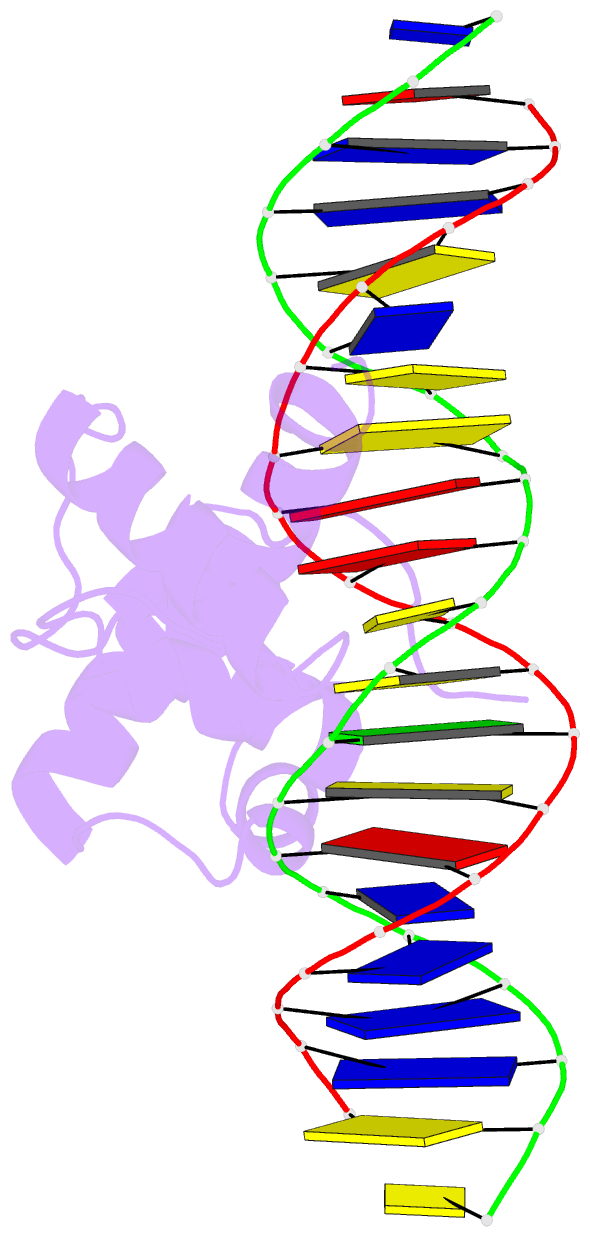
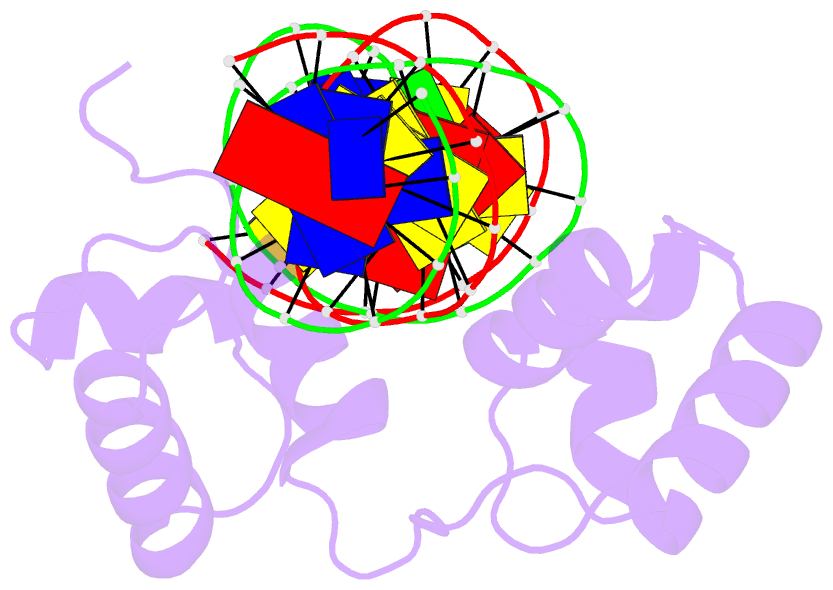
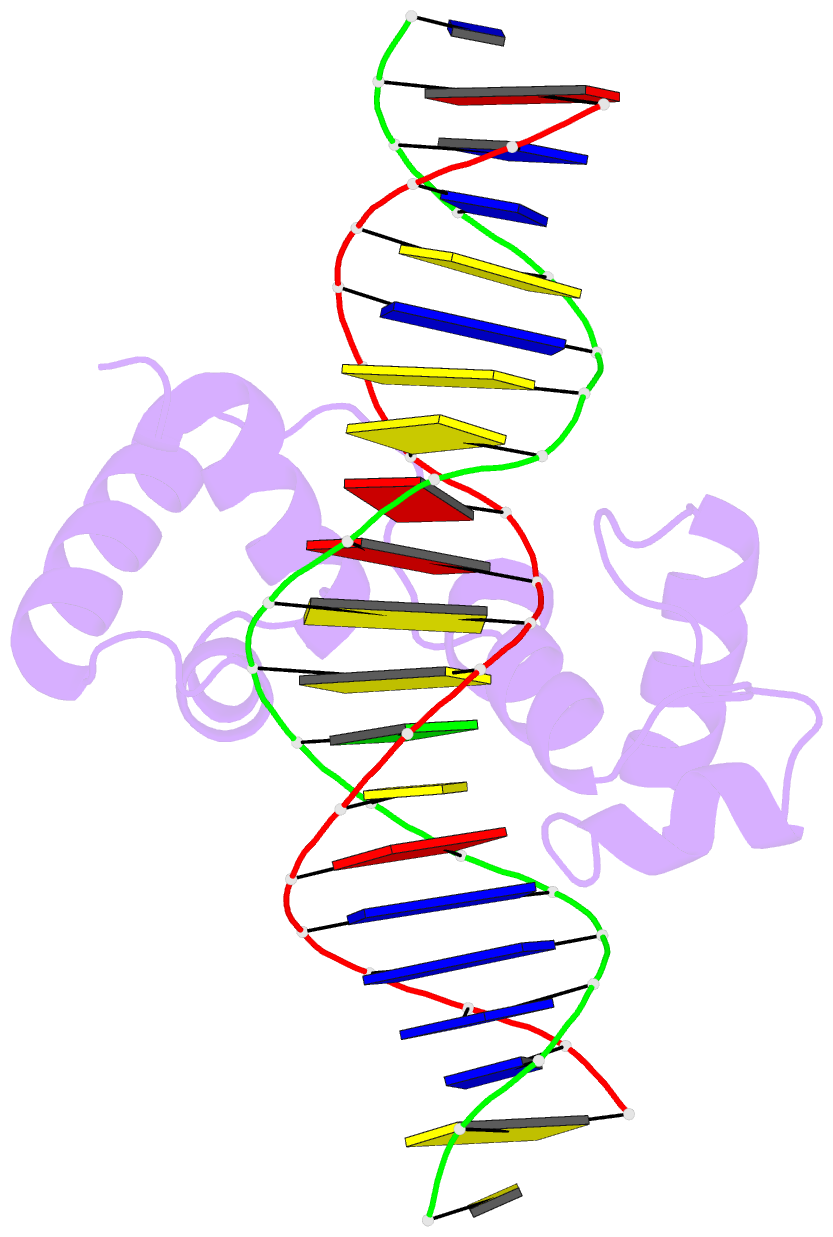
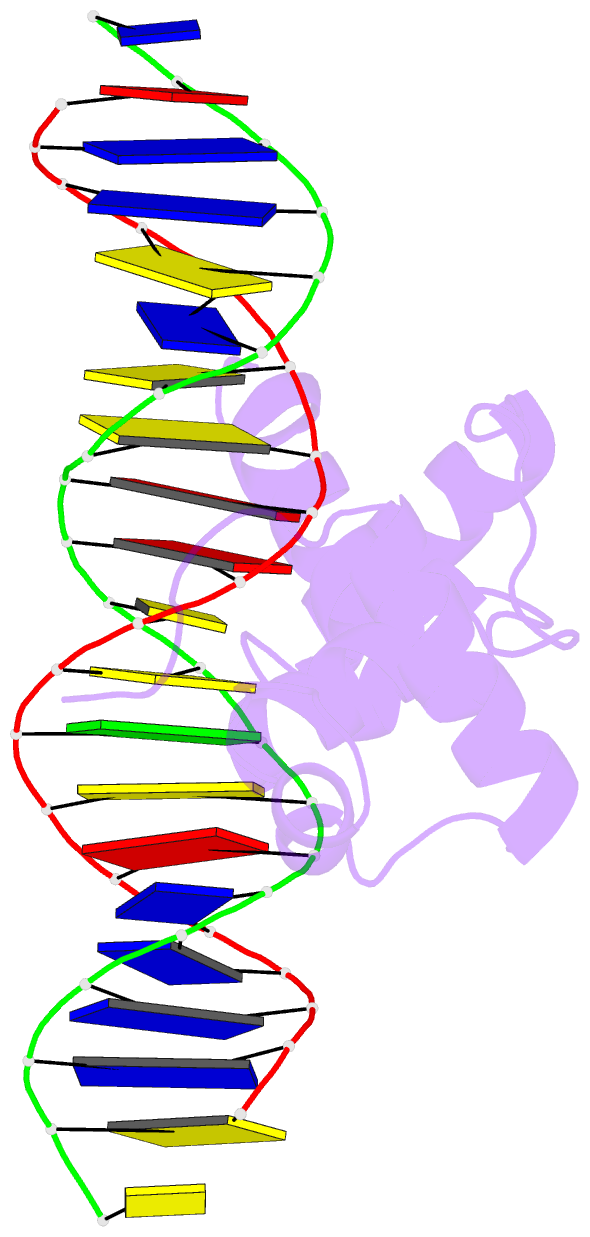
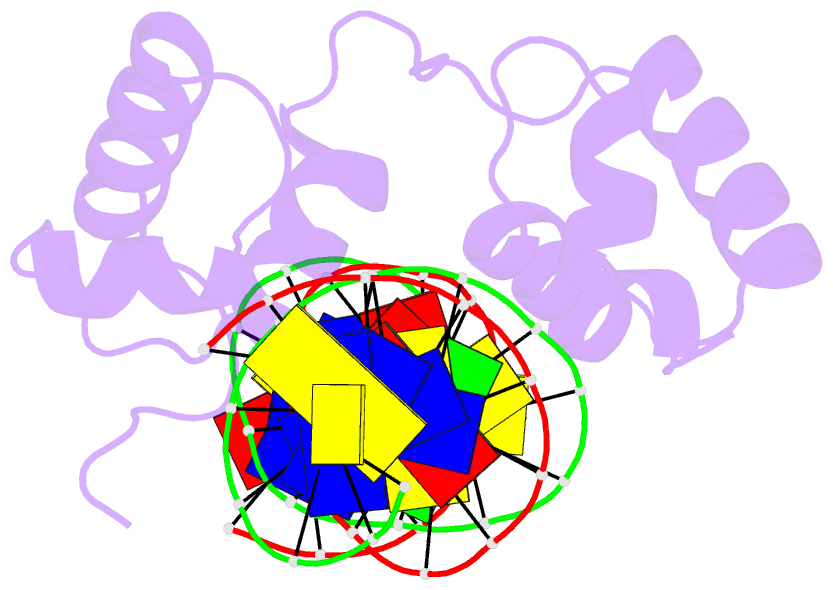
- Structural insights into target DNA recognition by r2r3-type myb transcription factor
- Reference
- Wang B, Luo Q, Li Y, Yin L, Zhou N, Li X, Gan J, Dong A (2020): "Structural insights into target DNA recognition by R2R3-MYB transcription factors." Nucleic Acids Res., 48, 460-471. doi: 10.1093/nar/gkz1081.
- Abstract
- As the largest group of MYB family transcription factors, R2R3-MYB proteins play essential roles during plant growth and development. However, the structural basis underlying how R2R3-MYBs recognize the target DNA remains elusive. Here, we report the crystal structure of Arabidopsis WEREWOLF (WER), an R2R3-MYB protein, in complex with its target DNA. Structural analysis showed that the third α-helices in both the R2 and R3 repeats of WER fit in the major groove of the DNA, specifically recognizing the DNA motif 5'-AACNGC-3'. In combination with mutagenesis, in vitro binding and in vivo luciferase assays, we showed that K55, N106, K109 and N110 are critical for the function of WER. Although L59 of WER is not involved in DNA binding in the structure, ITC analysis suggested that L59 plays an important role in sensing DNA methylation at the fifth position of cytosine (5mC). Like 5mC, methylation at the sixth position of adenine (6mA) in the AAC element also inhibits the interaction between WER and its target DNA. Our study not only unravels the molecular basis of how WER recognizes its target DNA, but also suggests that 5mC and 6mA modifications may block the interaction between R2R3-MYB transcription factors and their target genes.