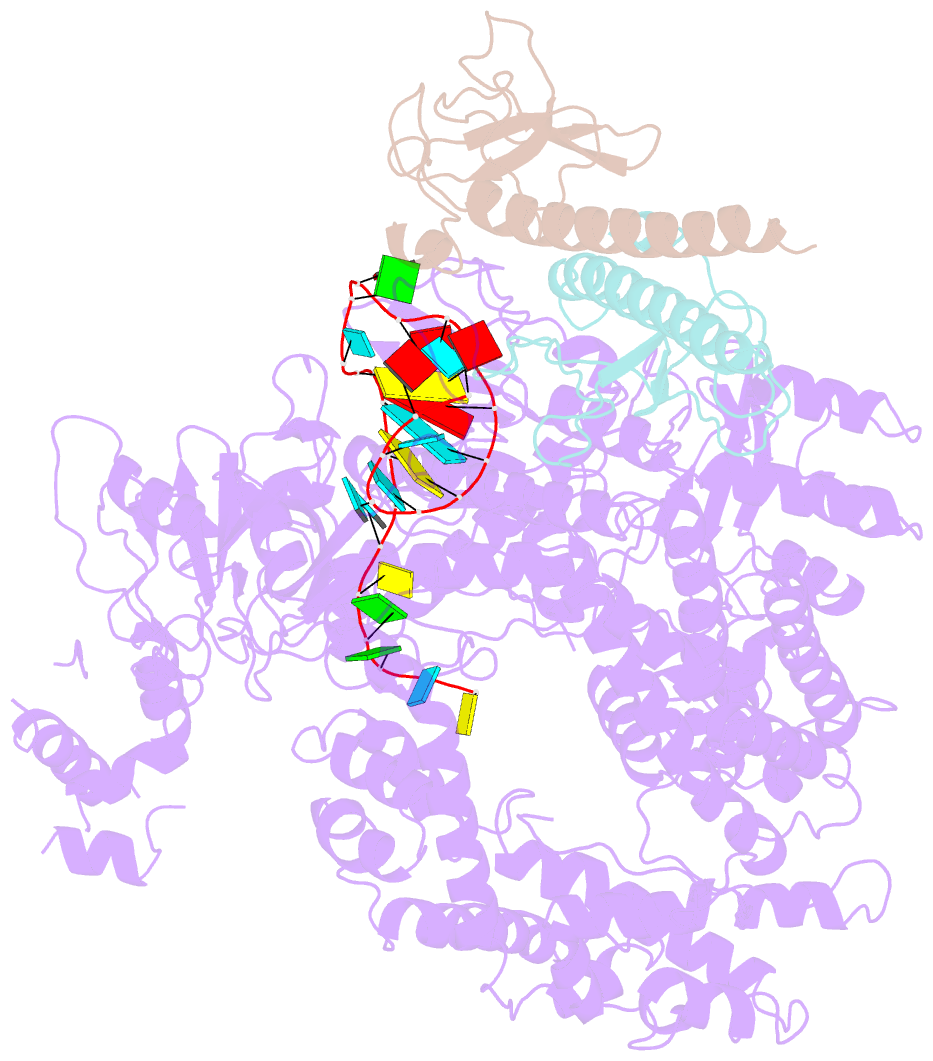
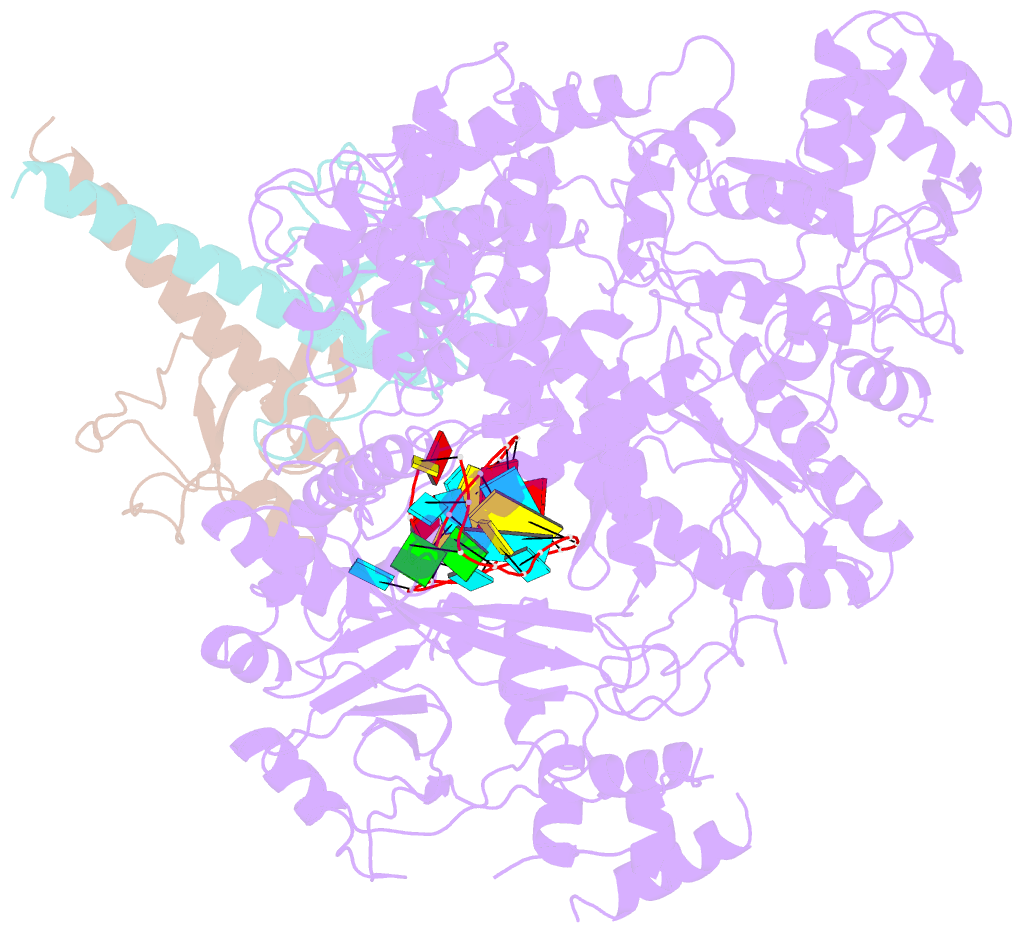
Summary information and primary citation
- PDB-id
- 6kl9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.25 Å)
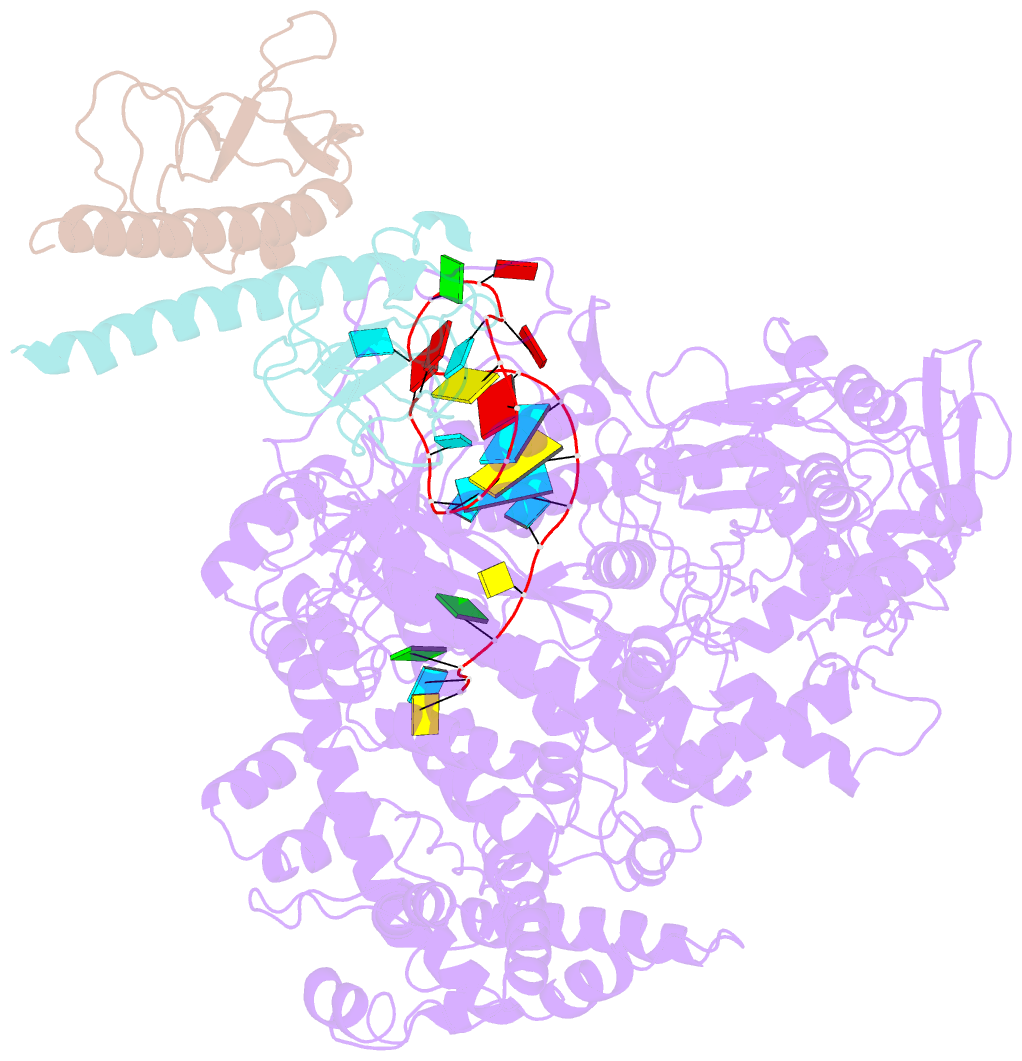
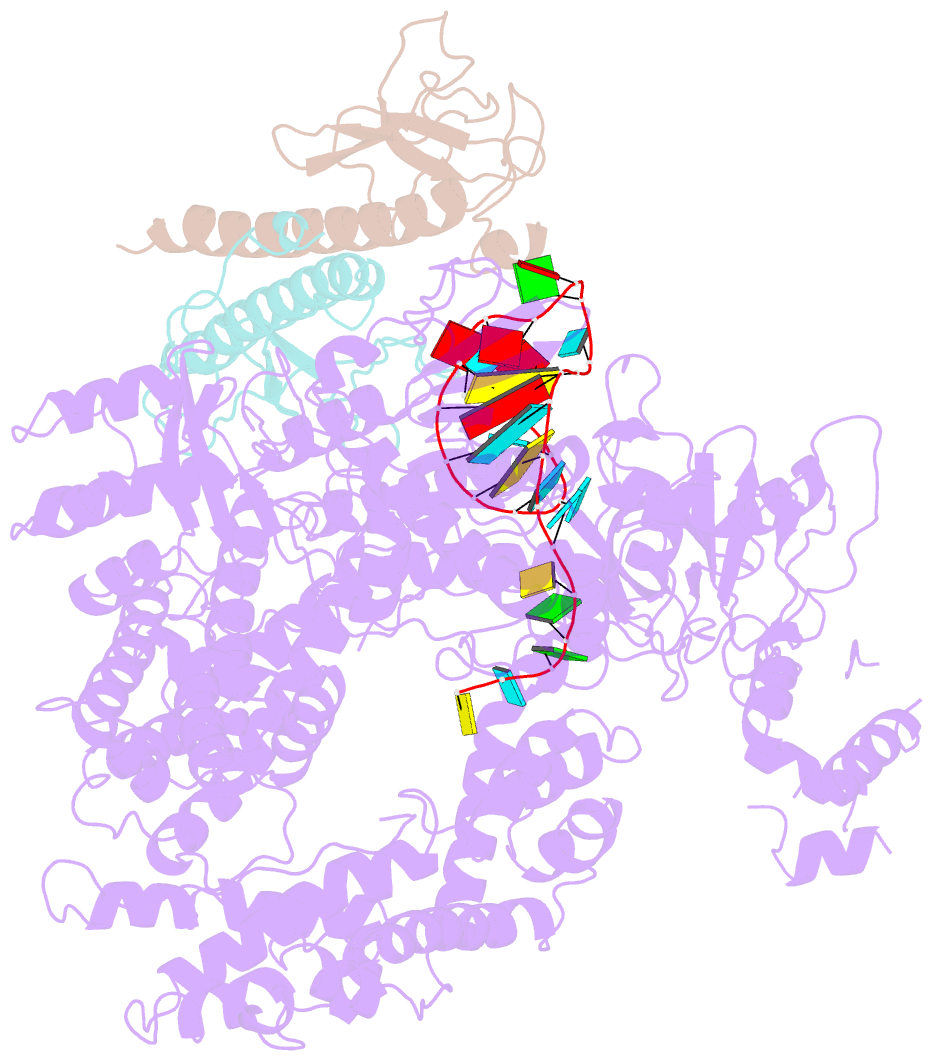
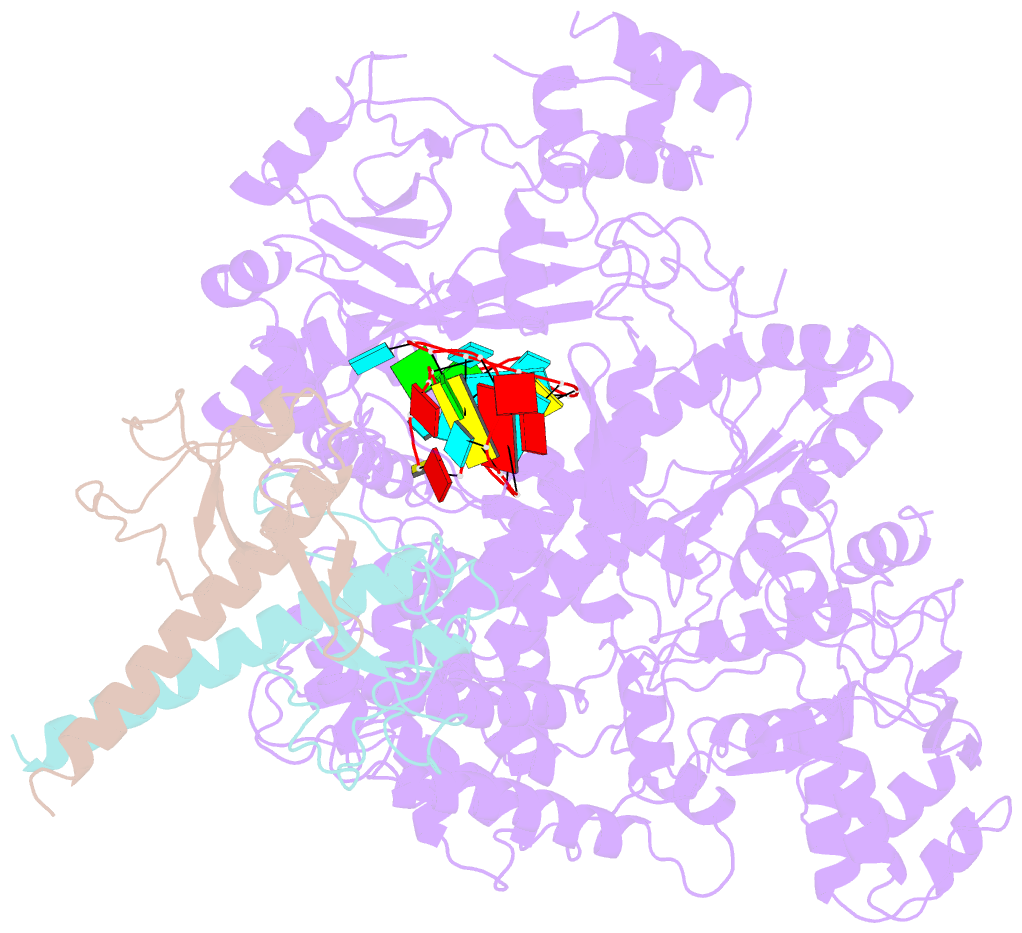
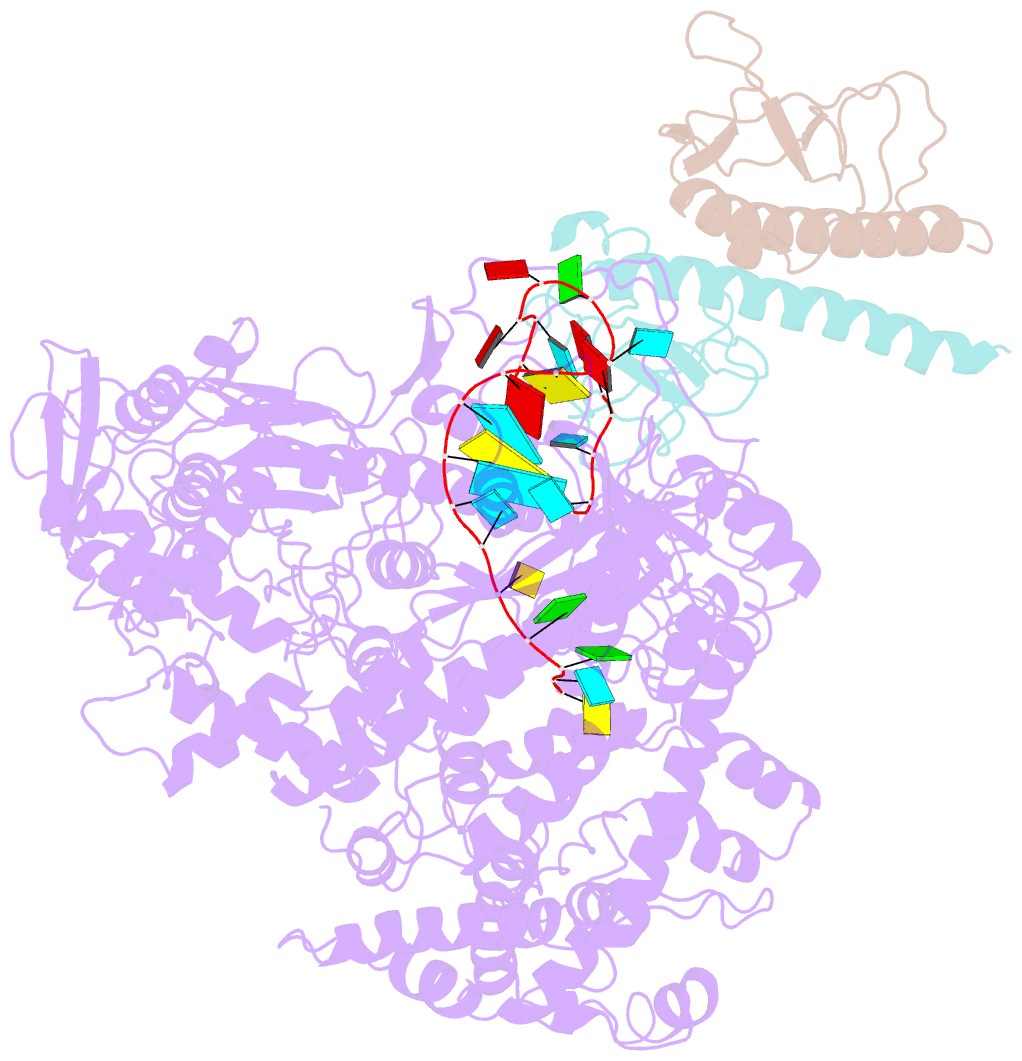
- Summary
- Structure of lbcas12a-crrna complex bound to acrva4 (form a complex)
- Reference
- Peng R, Li Z, Xu Y, He S, Peng Q, Wu LA, Wu Y, Qi J, Wang P, Shi Y, Gao GF (2019): "Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4." Proc.Natl.Acad.Sci.USA, 116, 18928-18936. doi: 10.1073/pnas.1909400116.
- Abstract
- Prokaryotes possess CRISPR-Cas systems to exclude parasitic predators, such as phages and mobile genetic elements (MGEs). These predators, in turn, encode anti-CRISPR (Acr) proteins to evade the CRISPR-Cas immunity. Recently, AcrVA4, an Acr protein inhibiting the CRISPR-Cas12a system, was shown to diminish Lachnospiraceae bacterium Cas12a (LbCas12a)-mediated genome editing in human cells, but the underlying mechanisms remain elusive. Here we report the cryo-EM structures of AcrVA4 bound to CRISPR RNA (crRNA)-loaded LbCas12a and found AcrVA4 could inhibit LbCas12a at several stages of the CRISPR-Cas working pathway, different from other characterized type I/II Acr inhibitors which target only 1 stage. First, it locks the conformation of the LbCas12a-crRNA complex to prevent target DNA-crRNA hybridization. Second, it interacts with the LbCas12a-crRNA-dsDNA complex to release the bound DNA before cleavage. Third, AcrVA4 binds the postcleavage LbCas12a complex to possibly block enzyme recycling. These findings highlight the multifunctionality of AcrVA4 and provide clues for developing regulatory genome-editing tools.