Summary information and primary citation
- PDB-id
- 6kw3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- cryo-EM (7.13 Å)
- Summary
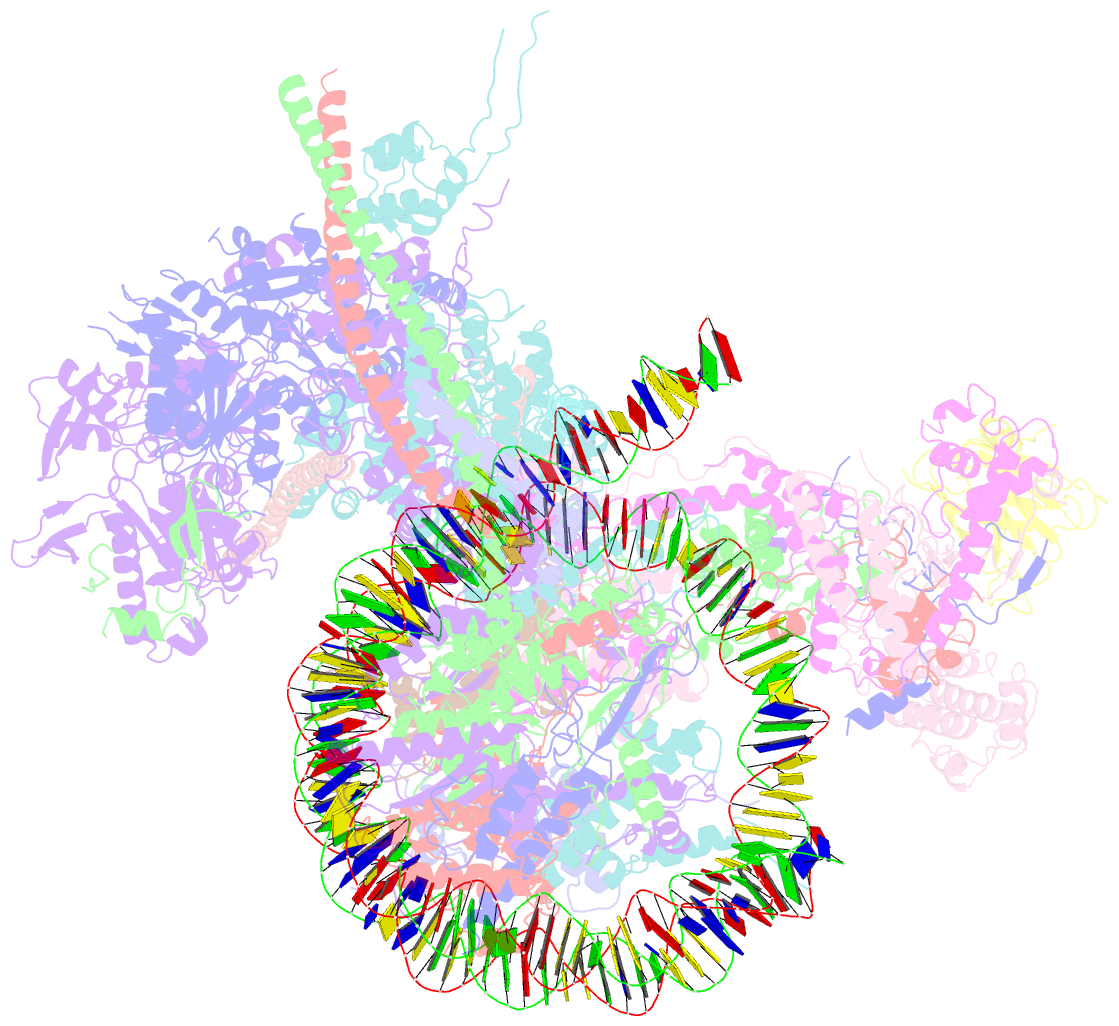
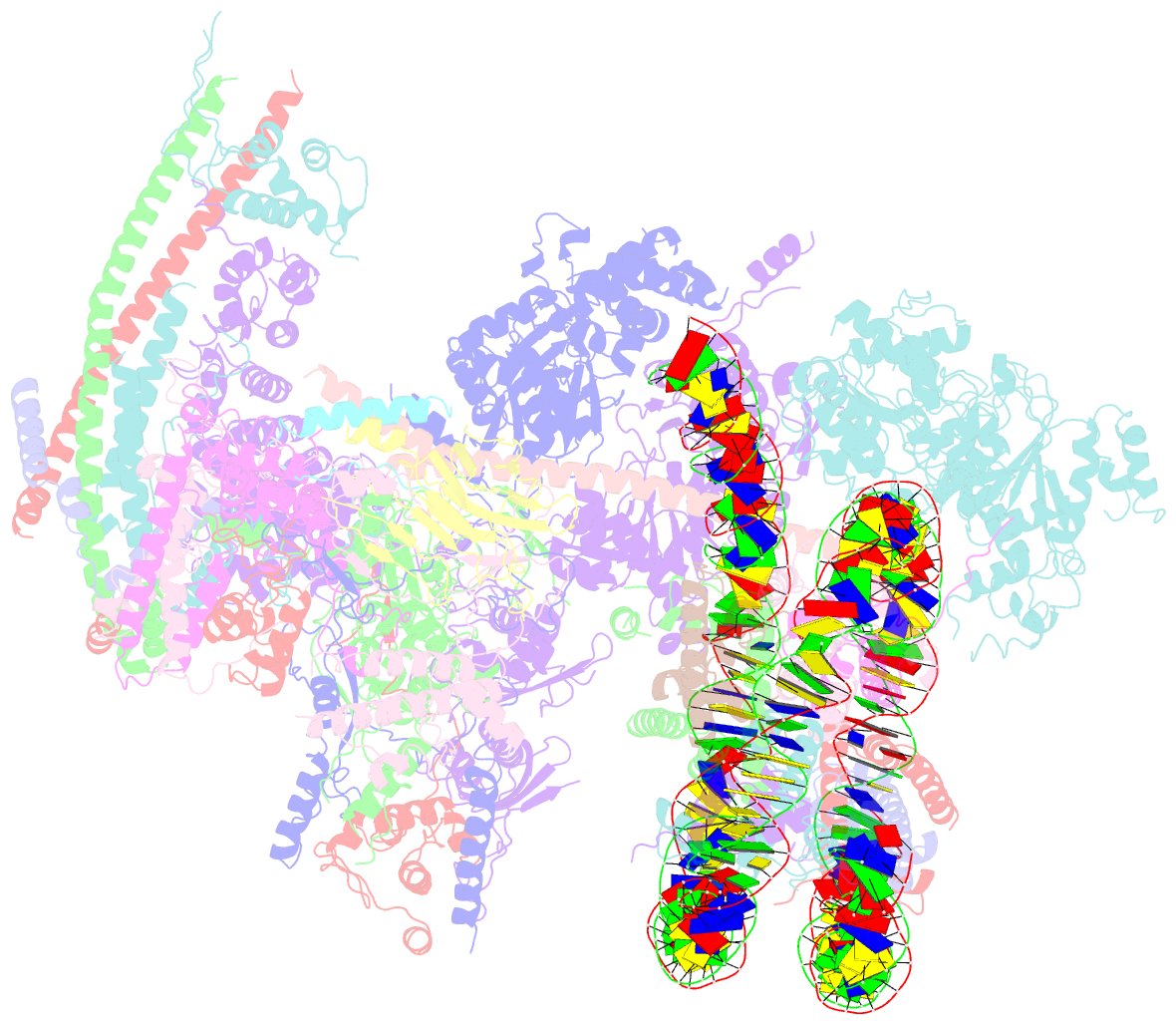
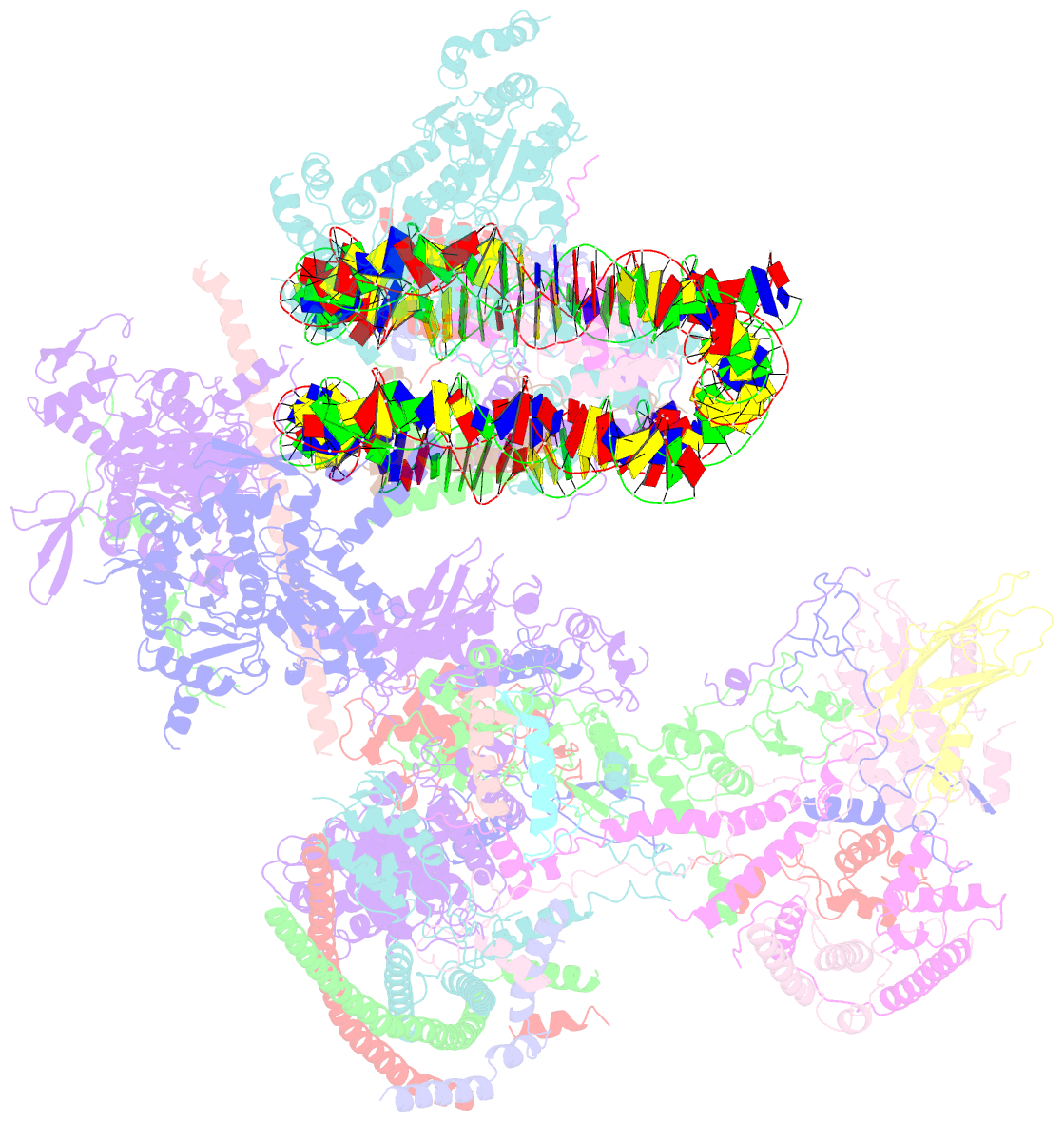
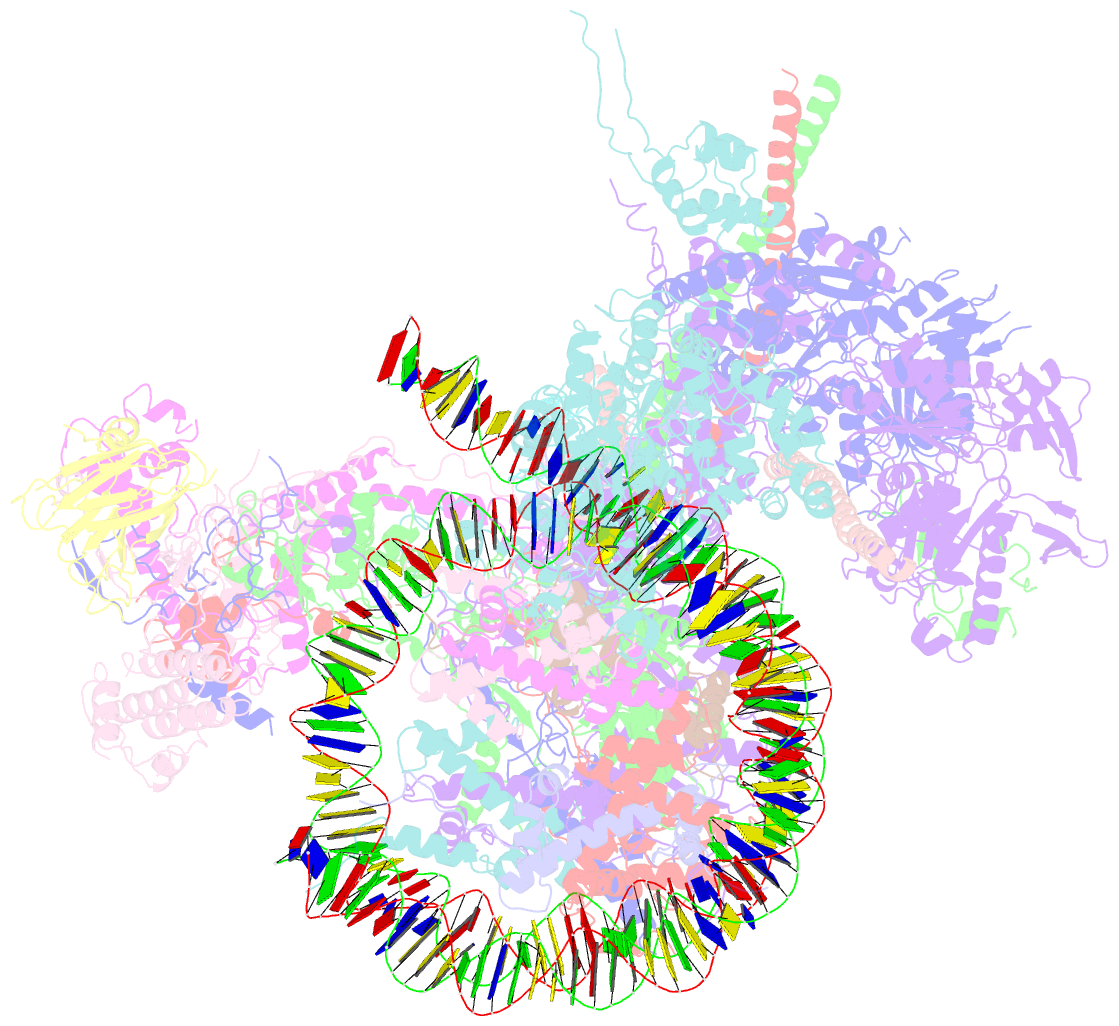
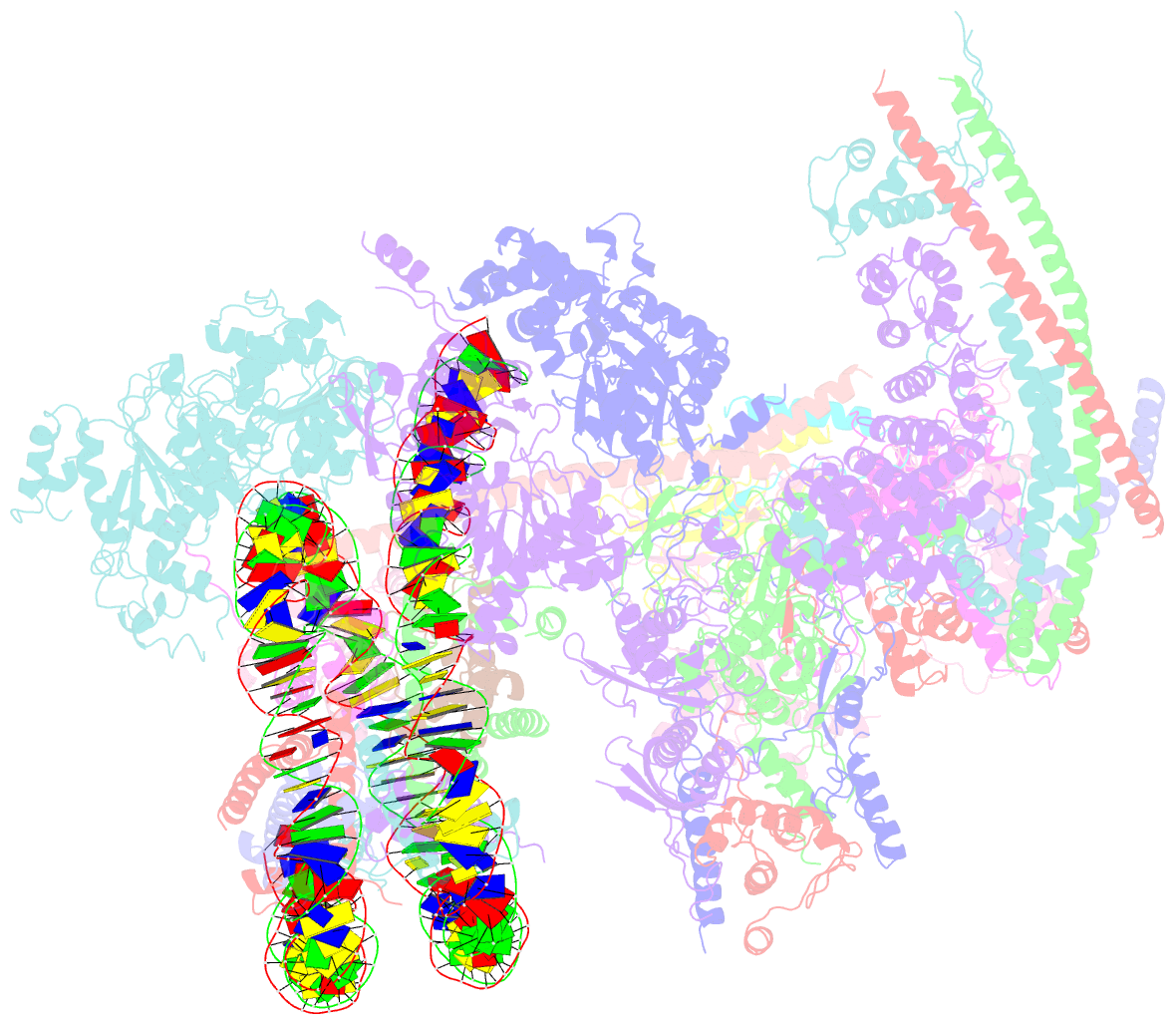
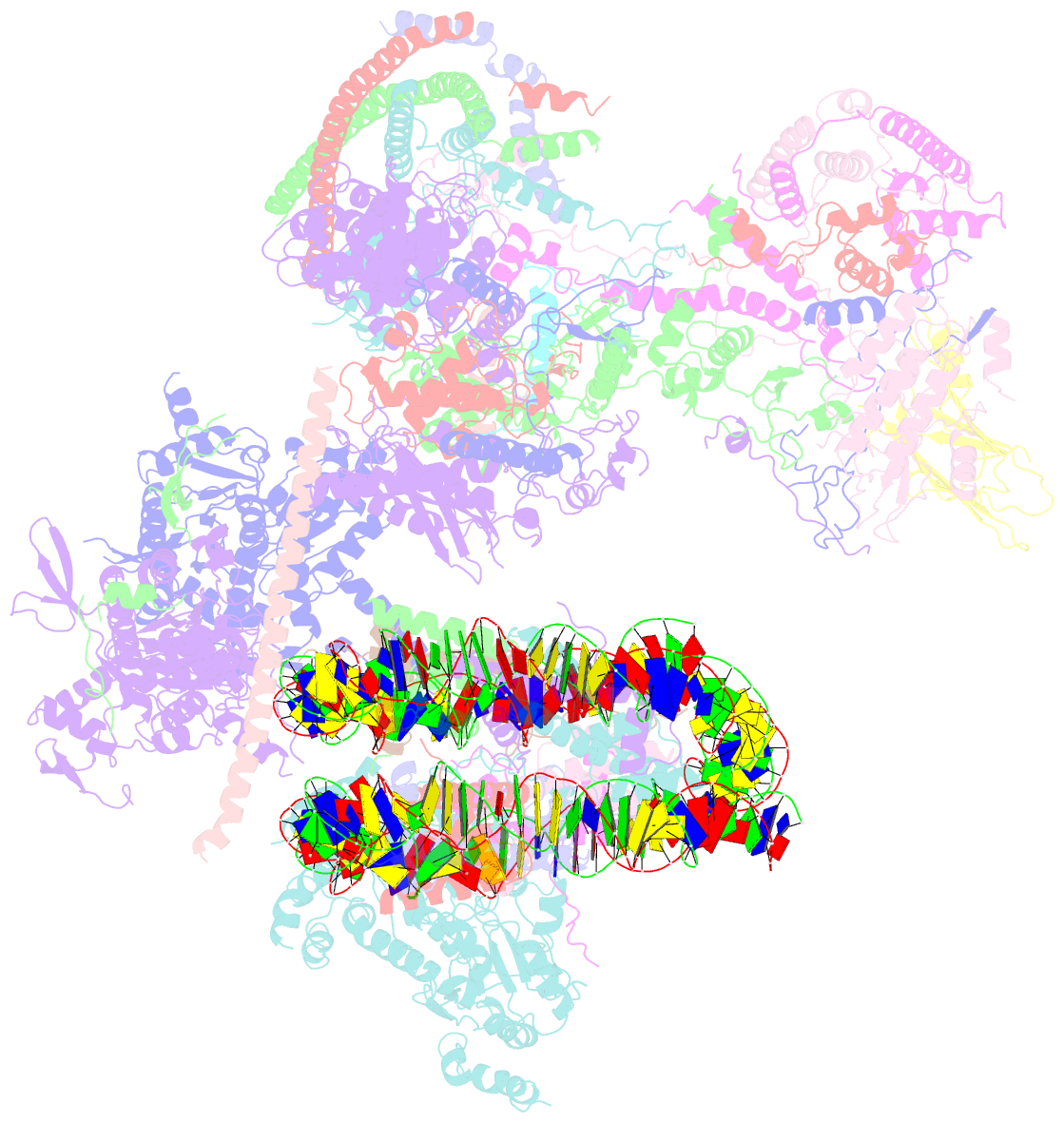
- The classa rsc-nucleosome complex
- Reference
- Ye Y, Wu H, Chen K, Clapier CR, Verma N, Zhang W, Deng H, Cairns BR, Gao N, Chen Z (2019): "Structure of the RSC complex bound to the nucleosome." Science, 366, 838-843. doi: 10.1126/science.aay0033.
- Abstract
- The RSC complex remodels chromatin structure and regulates gene transcription. We used cryo-electron microscopy to determine the structure of yeast RSC bound to the nucleosome. RSC is delineated into the adenosine triphosphatase motor, the actin-related protein module, and the substrate recruitment module (SRM). RSC binds the nucleosome mainly through the motor, with the auxiliary subunit Sfh1 engaging the H2A-H2B acidic patch to enable nucleosome ejection. SRM is organized into three substrate-binding lobes poised to bind their respective nucleosomal epitopes. The relative orientations of the SRM and the motor on the nucleosome explain the directionality of DNA translocation and promoter nucleosome repositioning by RSC. Our findings shed light on RSC assembly and functionality, and they provide a framework to understand the mammalian homologs BAF/PBAF and the Sfh1 ortholog INI1/BAF47, which are frequently mutated in cancers.