Summary information and primary citation
- PDB-id
- 6l9z; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.5 Å)
- Summary
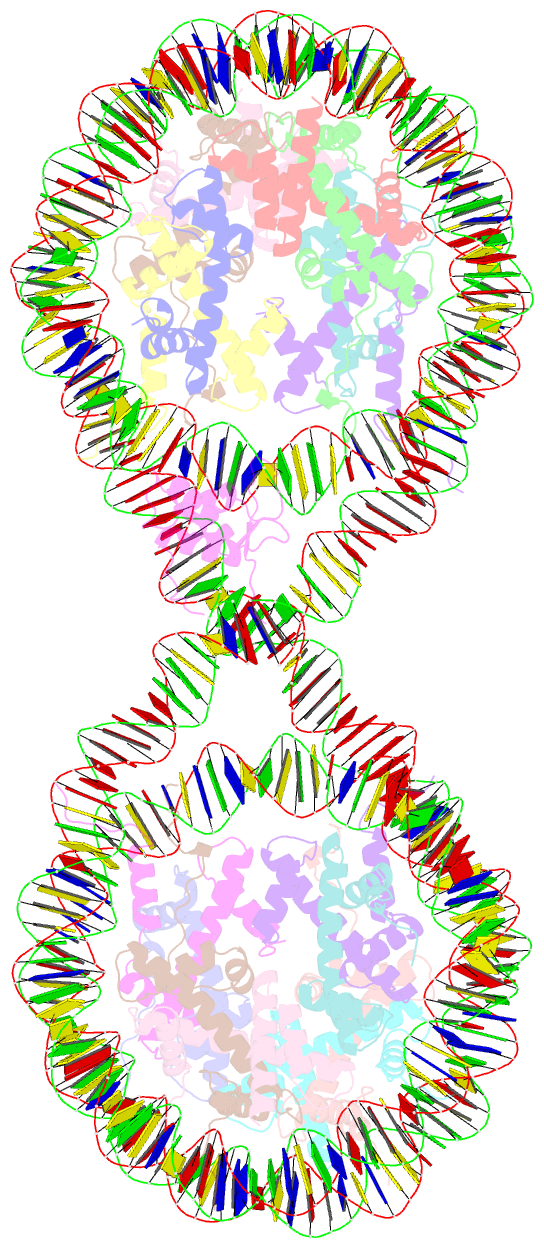
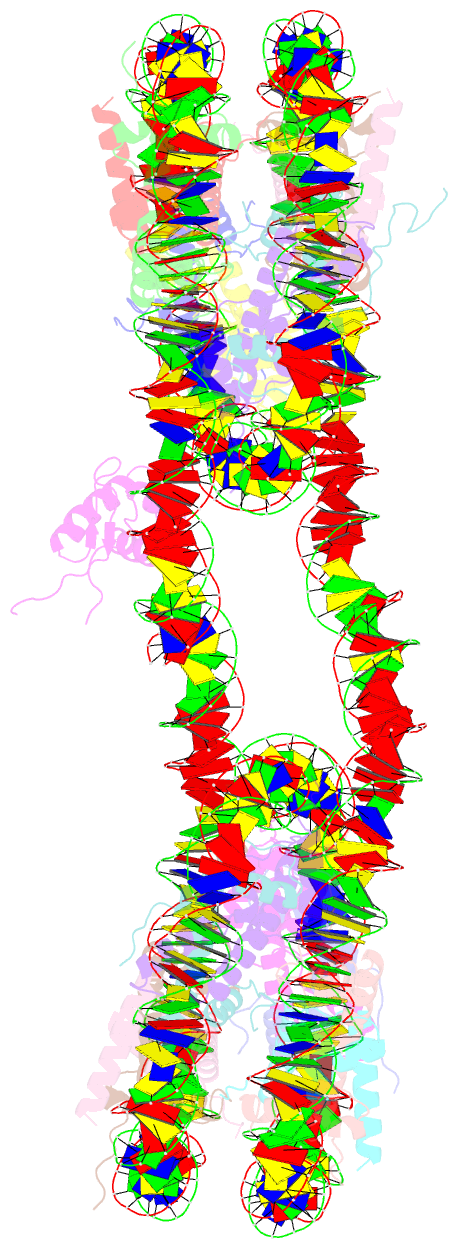
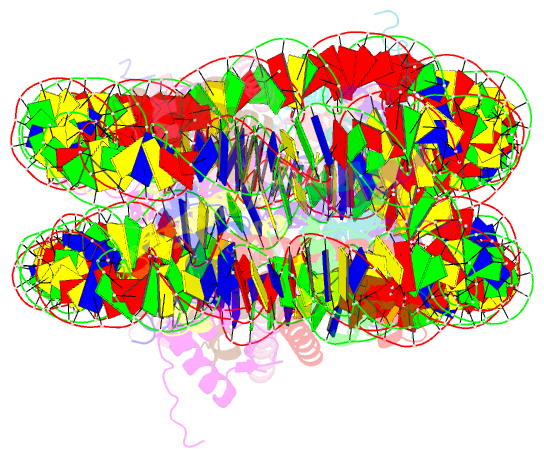
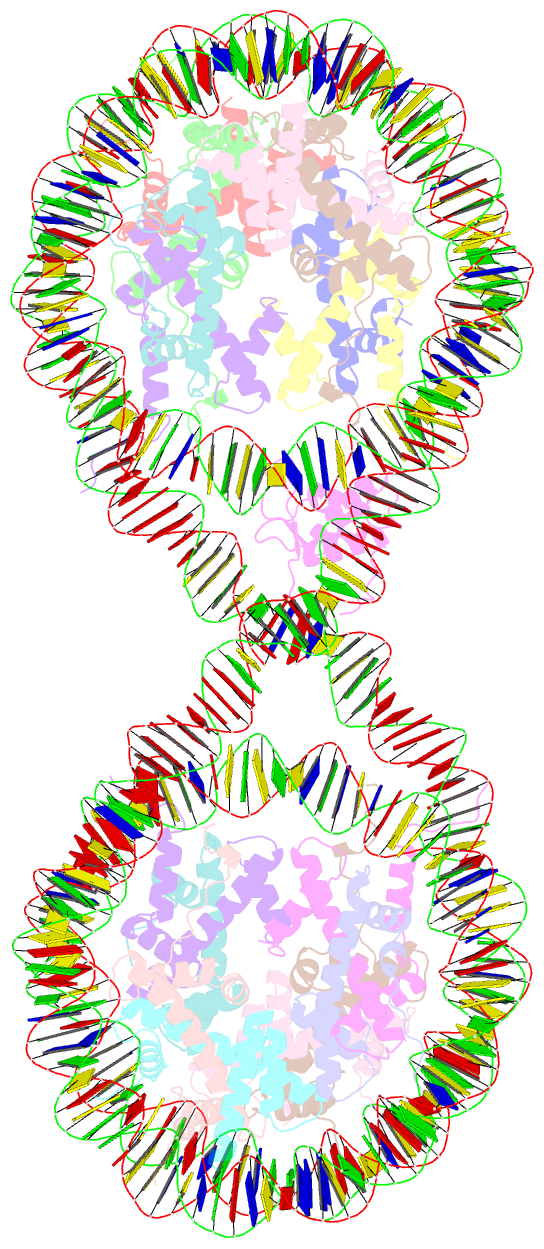
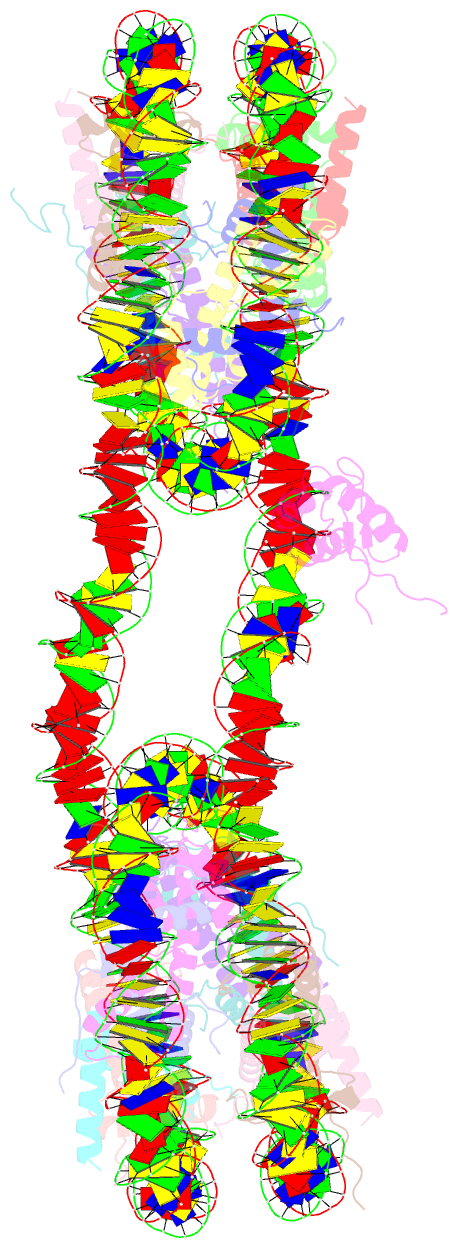
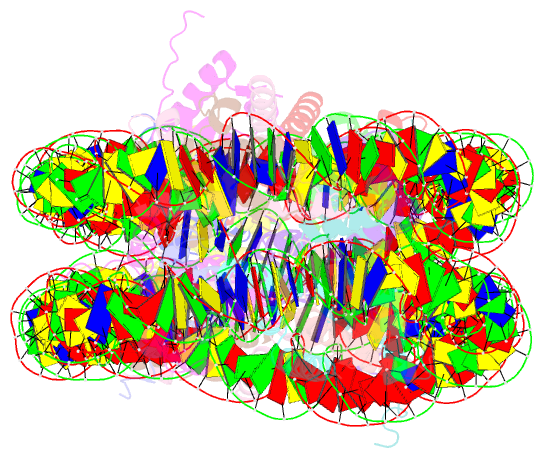
- 338 bp di-nucleosome assembled with linker histone h1.x
- Reference
- Adhireksan Z, Sharma D, Lee PL, Bao Q, Padavattan S, Shum WK, Davey GE, Davey CA (2021): "Engineering nucleosomes for generating diverse chromatin assemblies." Nucleic Acids Res., 49, e52. doi: 10.1093/nar/gkab070.
- Abstract
- Structural characterization of chromatin is challenging due to conformational and compositional heterogeneity in vivo and dynamic properties that limit achievable resolution in vitro. Although the maximum resolution for solving structures of large macromolecular assemblies by electron microscopy has recently undergone profound increases, X-ray crystallographic approaches may still offer advantages for certain systems. One such system is compact chromatin, wherein the crystalline state recapitulates the crowded molecular environment within the nucleus. Here we show that nucleosomal constructs with cohesive-ended DNA can be designed that assemble into different types of circular configurations or continuous fibers extending throughout crystals. We demonstrate the utility of the method for characterizing nucleosome compaction and linker histone binding at near-atomic resolution but also advance its application for tackling further problems in chromatin structural biology and for generating novel types of DNA nanostructures. We provide a library of cohesive-ended DNA fragment expression constructs and a strategy for engineering DNA-based nanomaterials with a seemingly vast potential variety of architectures and histone chemistries.