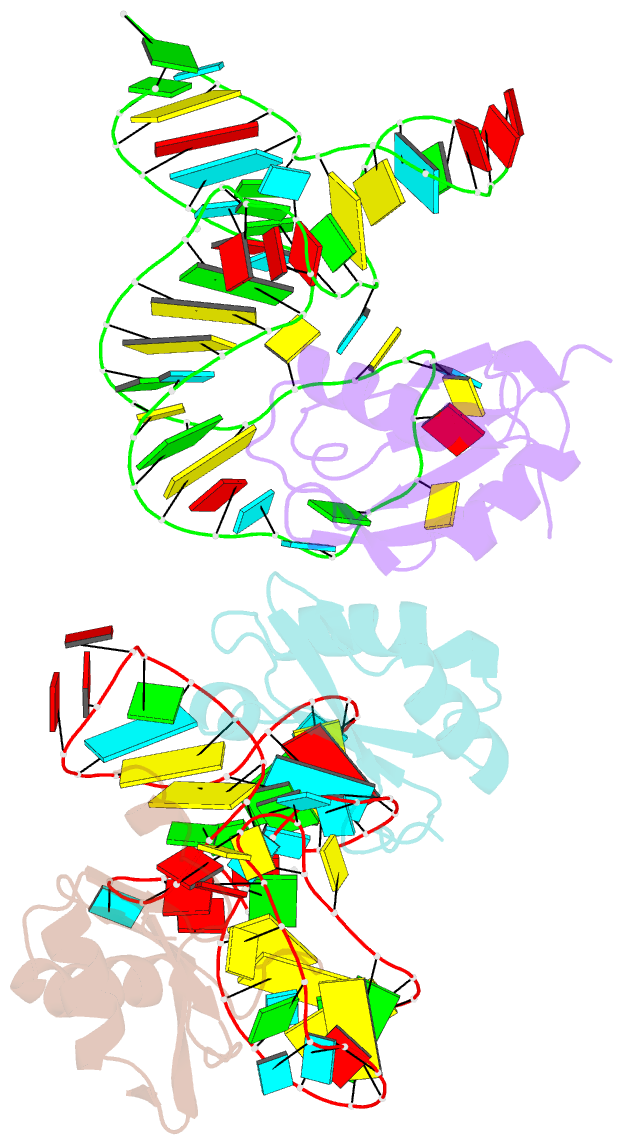
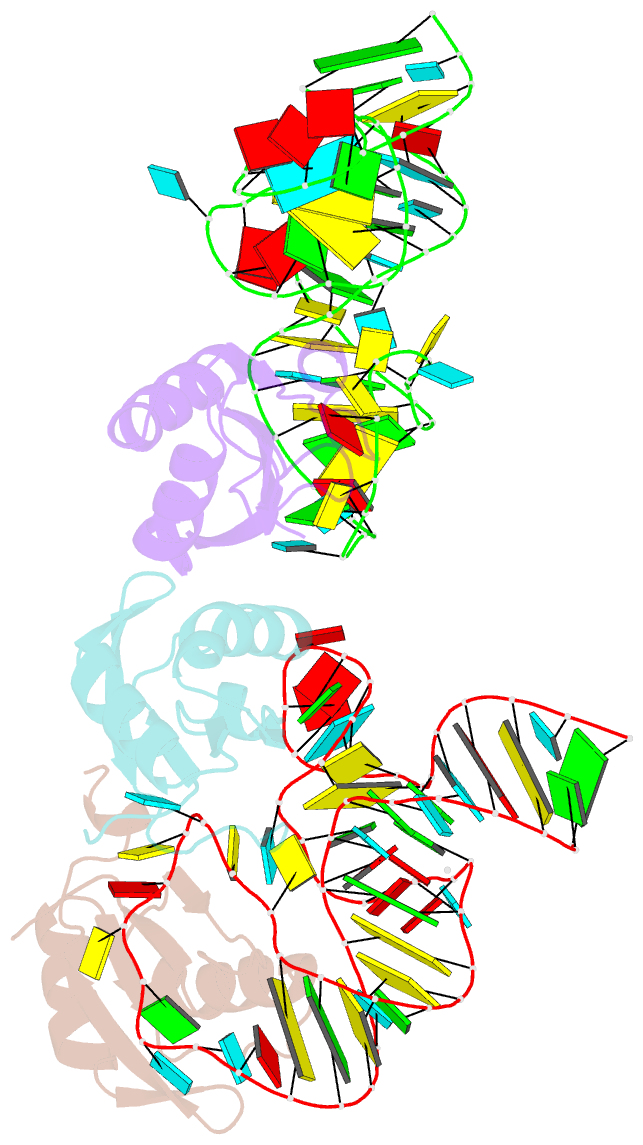
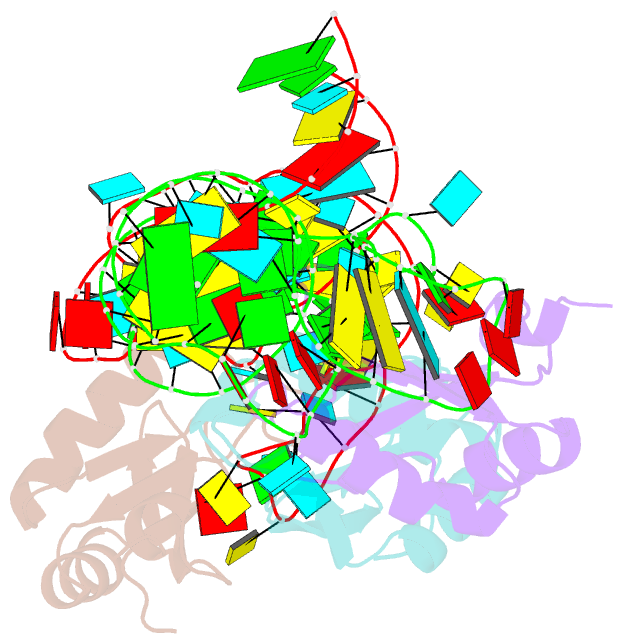
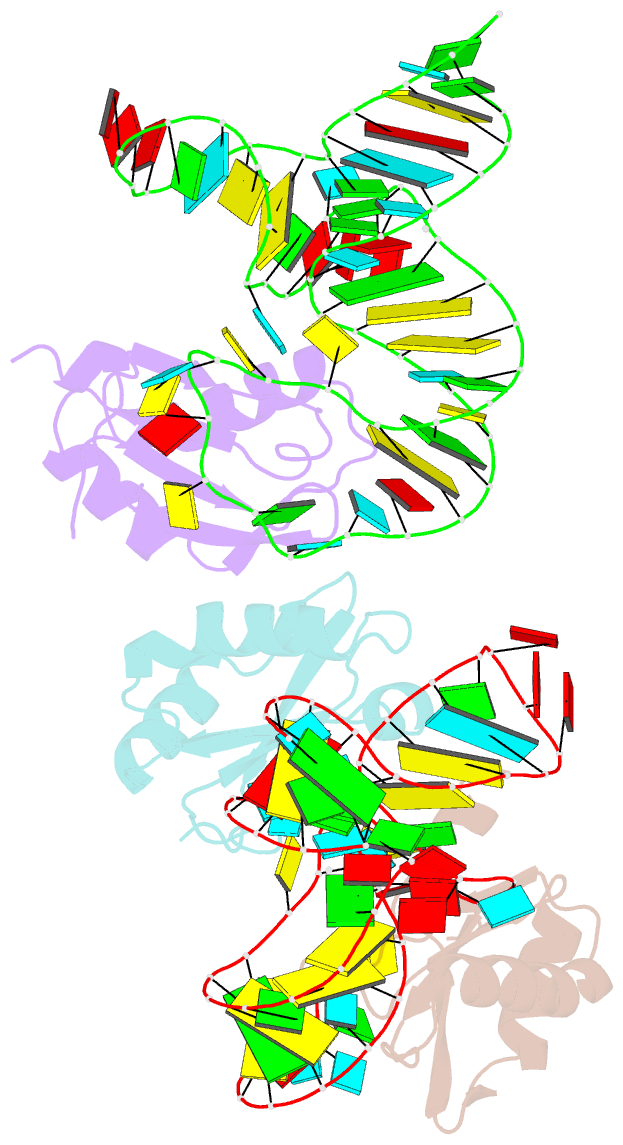
Summary information and primary citation
- PDB-id
- 6laz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA
- Method
- X-ray (2.76 Å)
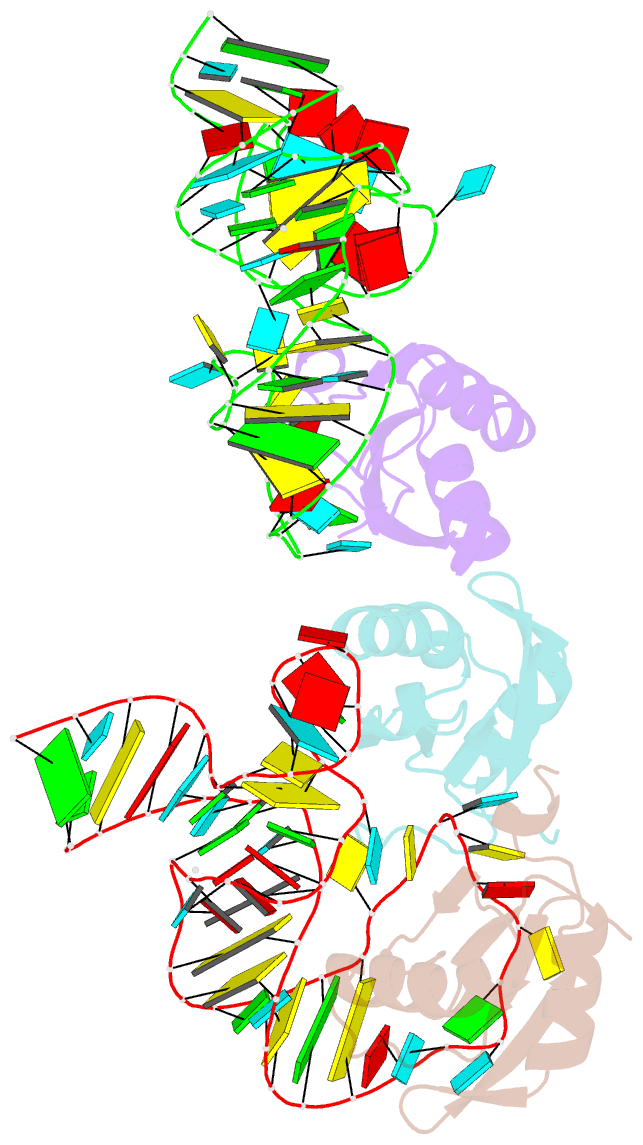
- Summary
- The wildtype sam-vi riboswitch bound to a n-mustard sam analog m1
- Reference
- Sun A, Gasser C, Li F, Chen H, Mair S, Krasheninina O, Micura R, Ren A (2019): "SAM-VI riboswitch structure and signature for ligand discrimination." Nat Commun, 10, 5728. doi: 10.1038/s41467-019-13600-9.
- Abstract
- Riboswitches are metabolite-sensing, conserved domains located in non-coding regions of mRNA that are central to regulation of gene expression. Here we report the first three-dimensional structure of the recently discovered S-adenosyl-L-methionine responsive SAM-VI riboswitch. SAM-VI adopts a unique fold and ligand pocket that are distinct from all other known SAM riboswitch classes. The ligand binds to the junctional region with its adenine tightly intercalated and Hoogsteen base-paired. Furthermore, we reveal the ligand discrimination mode of SAM-VI by additional X-ray structures of this riboswitch bound to S-adenosyl-L-homocysteine and a synthetic ligand mimic, in combination with isothermal titration calorimetry and fluorescence spectroscopy to explore binding thermodynamics and kinetics. The structure is further evaluated by analysis of ligand binding to SAM-VI mutants. It thus provides a thorough basis for developing synthetic SAM cofactors for applications in chemical and synthetic RNA biology.