Summary information and primary citation
- PDB-id
- 6lrd; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.9 Å)
- Summary
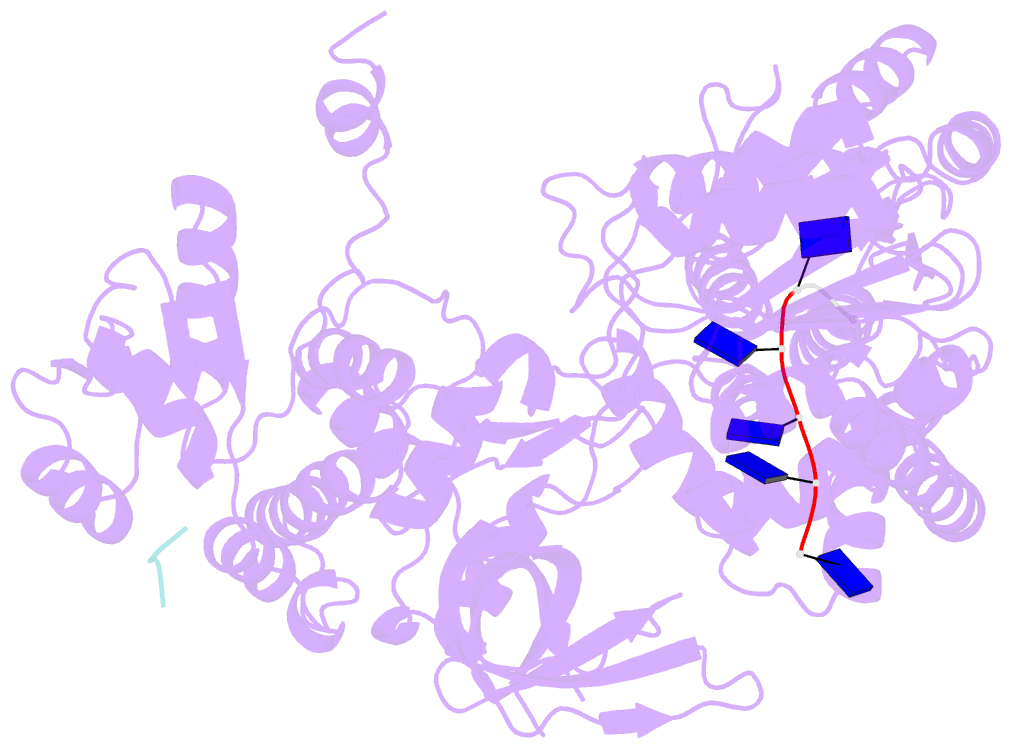
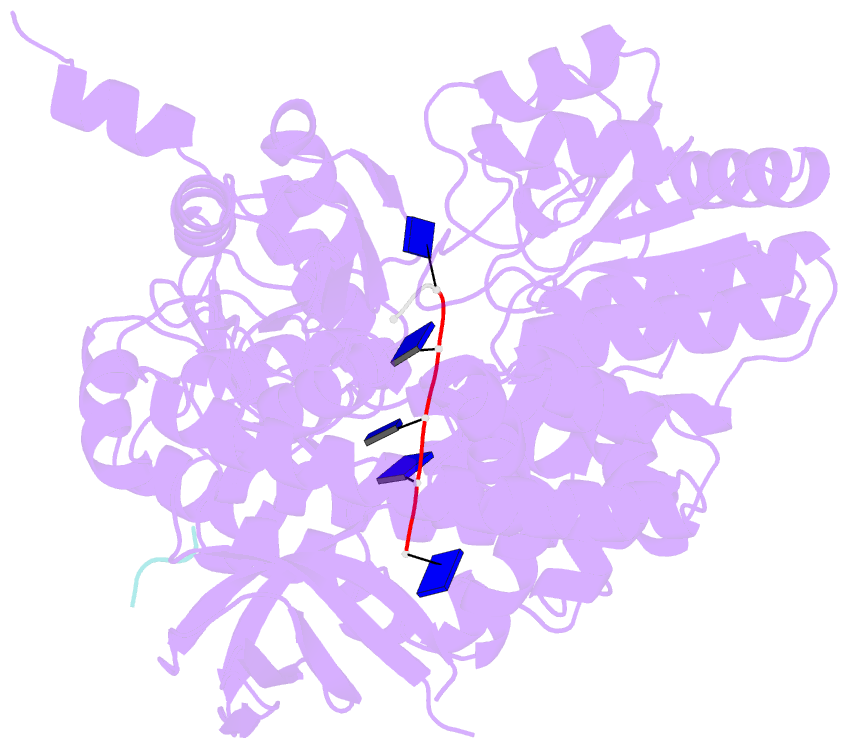
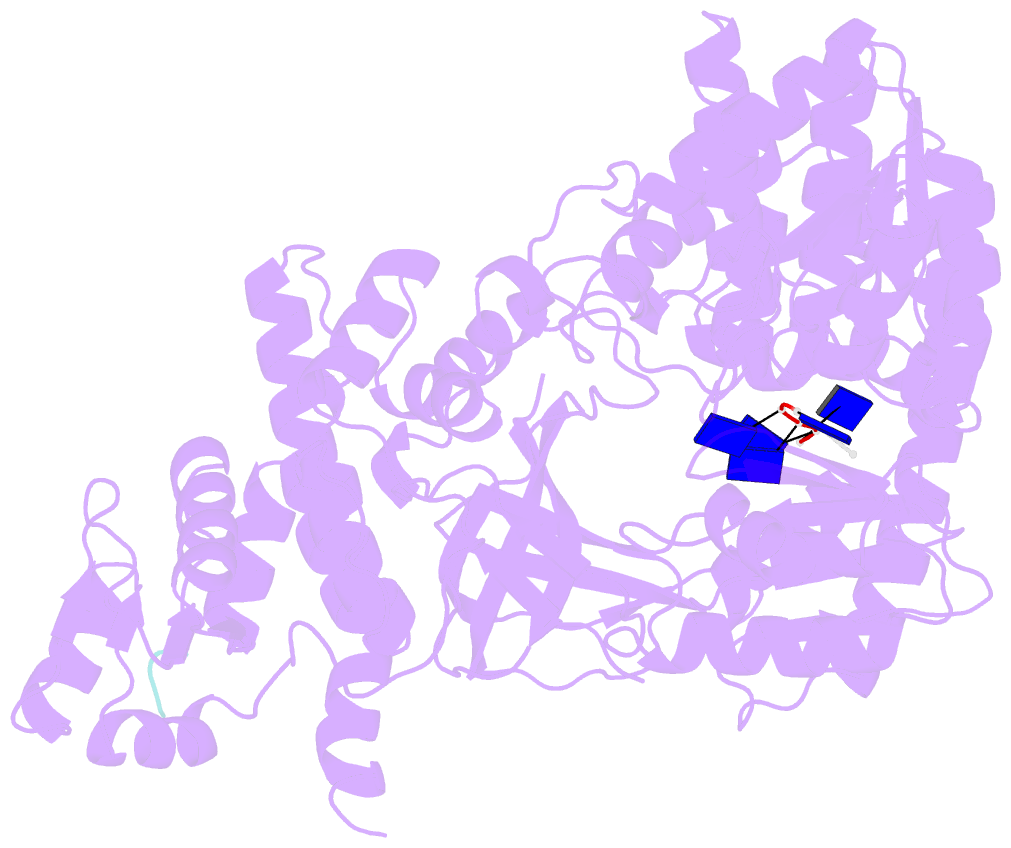
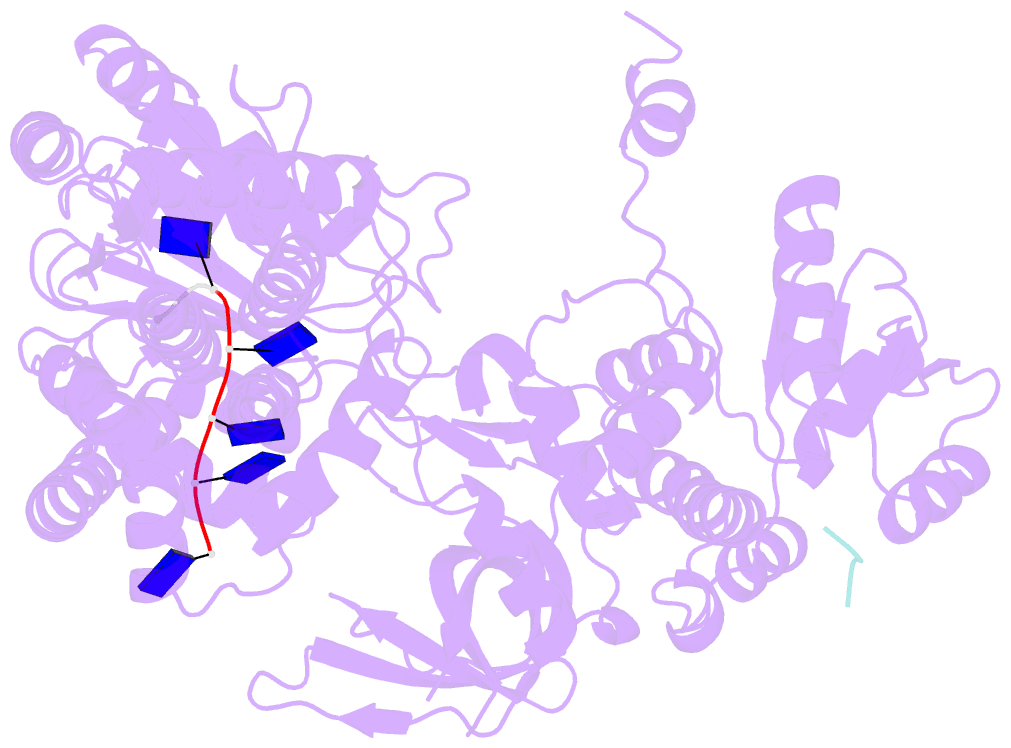
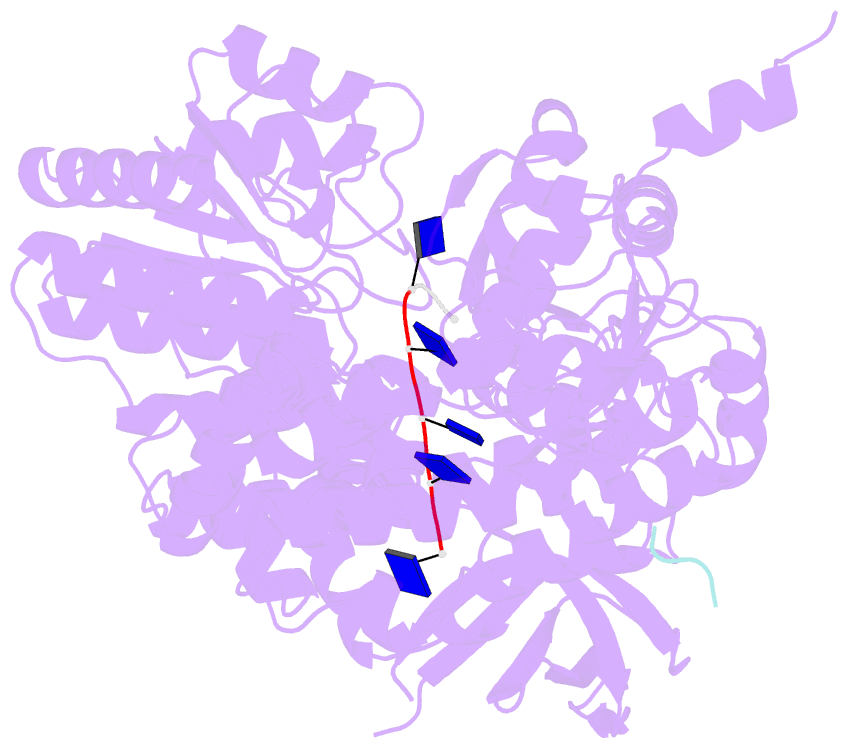
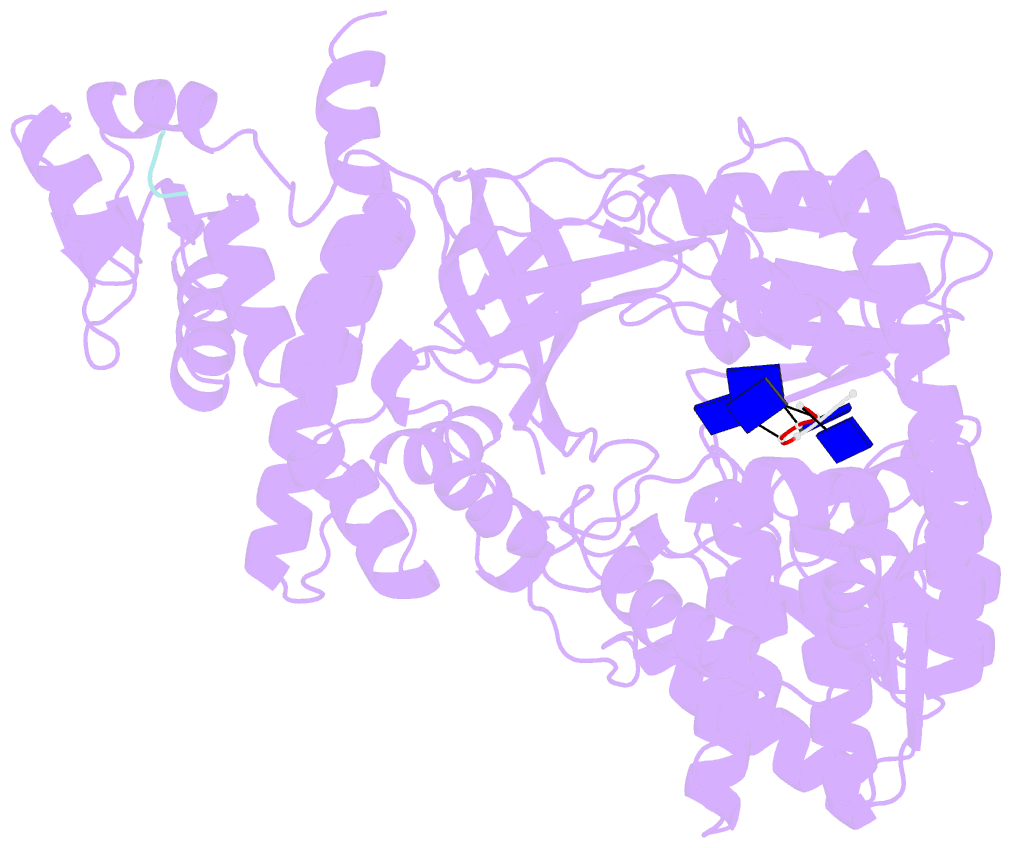
- Structure of recj complexed with a 5'-p-dspacer-modified ssDNA
- Reference
- Cheng K, Xu Y, Chen X, Lu H, He Y, Wang L, Hua Y (2020): "Participation of RecJ in the base excision repair pathway of Deinococcus radiodurans." Nucleic Acids Res., 48, 9859-9871. doi: 10.1093/nar/gkaa714.
- Abstract
- RecJ reportedly participates in the base excision repair (BER) pathway, but structural and functional data are scarce. Herein, the Deinococcus radiodurans RecJ (drRecJ) deletion strain exhibited extreme sensitivity to hydrogen peroxide and methyl-methanesulphonate, as well as a high spontaneous mutation rate and an accumulation of unrepaired abasic sites in vivo, indicating the involvement of drRecJ in the BER pathway. The binding affinity and nuclease activity preference of drRecJ toward DNA substrates containing a 5'-P-dSpacer group, a 5'-deoxyribose-phosphate (dRP) mimic, were established. A 1.9 Å structure of drRecJ in complex with 5'-P-dSpacer-modified single-stranded DNA (ssDNA) revealed a 5'-monophosphate binding pocket and occupancy of 5'-dRP in the drRecJ nuclease core. The mechanism for RecJ 5'-dRP catalysis was explored using structural and biochemical data, and the results implied that drRecJ is not a canonical 5'-dRP lyase. Furthermore, in vitro reconstitution assays indicated that drRecJ tends to participate in the long-patch BER pathway rather than the short-patch BER pathway.