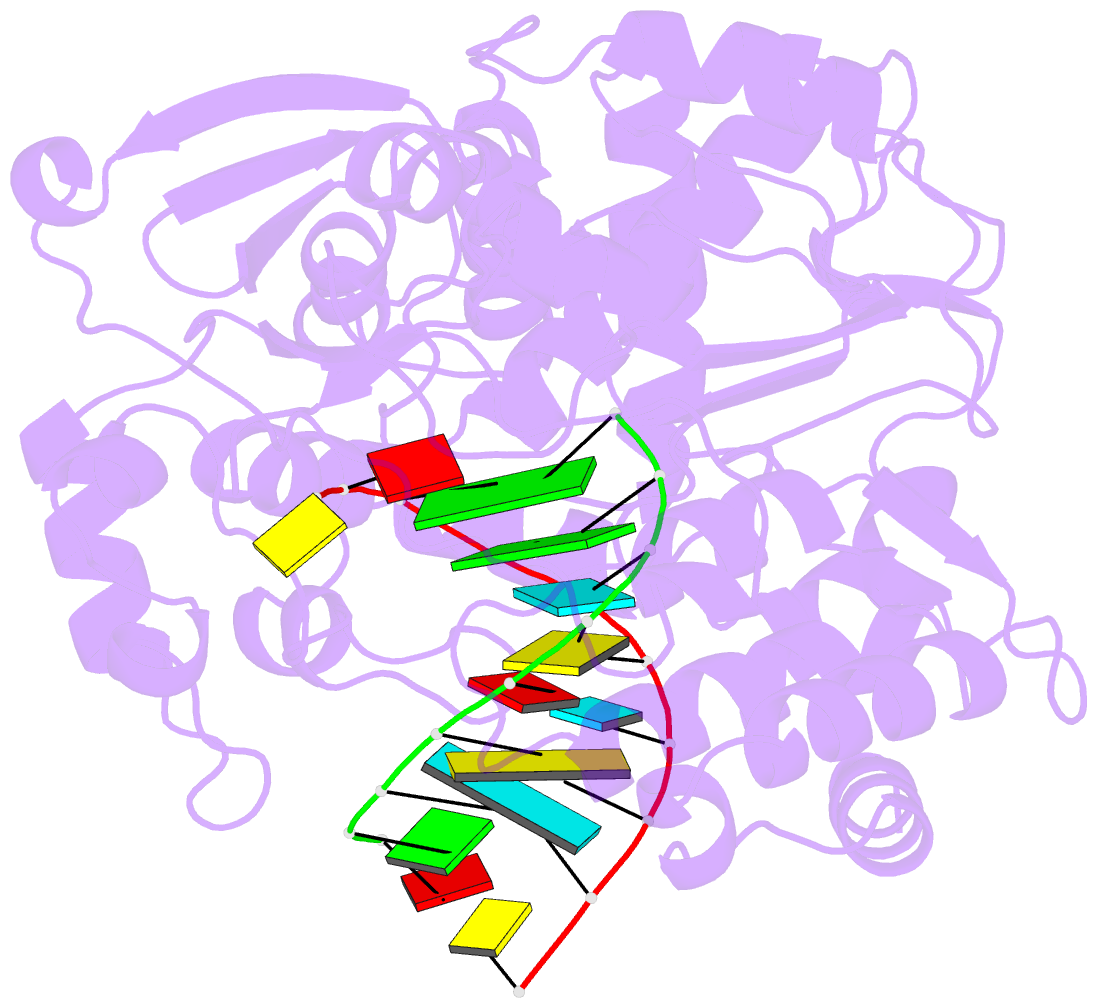
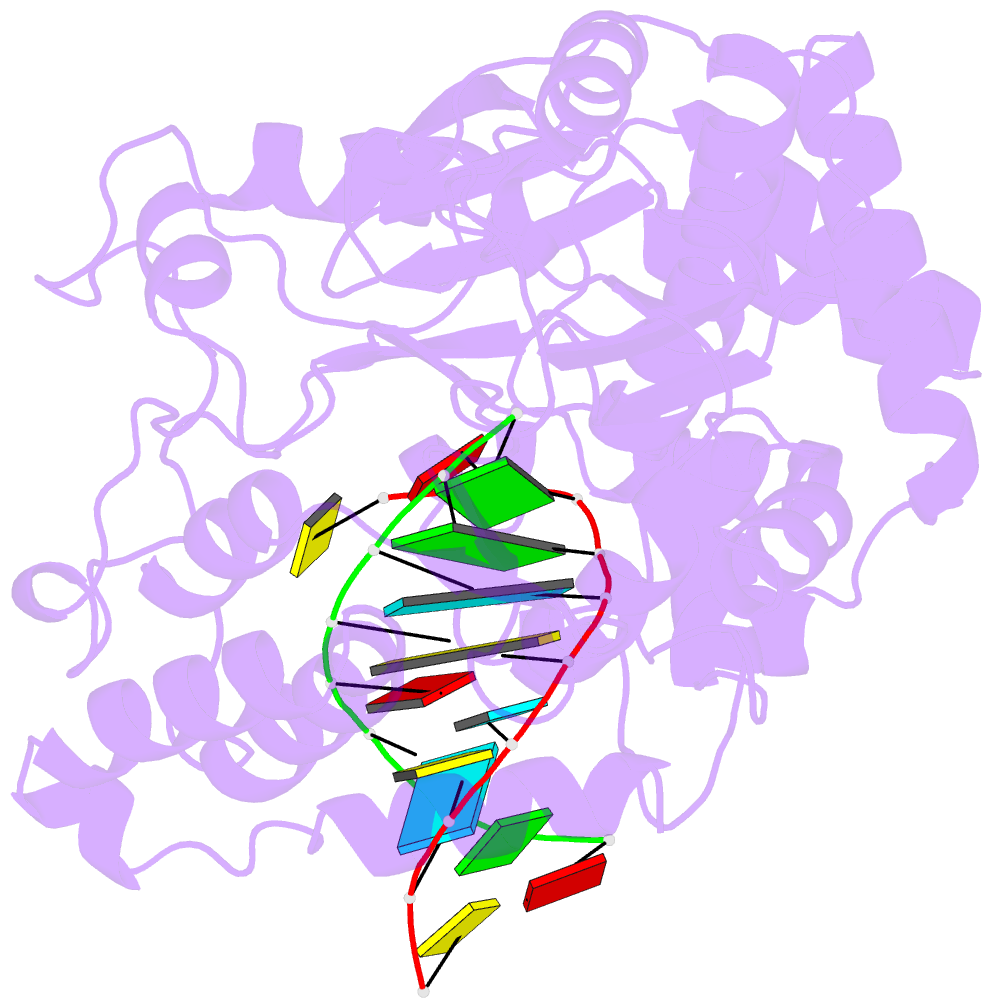
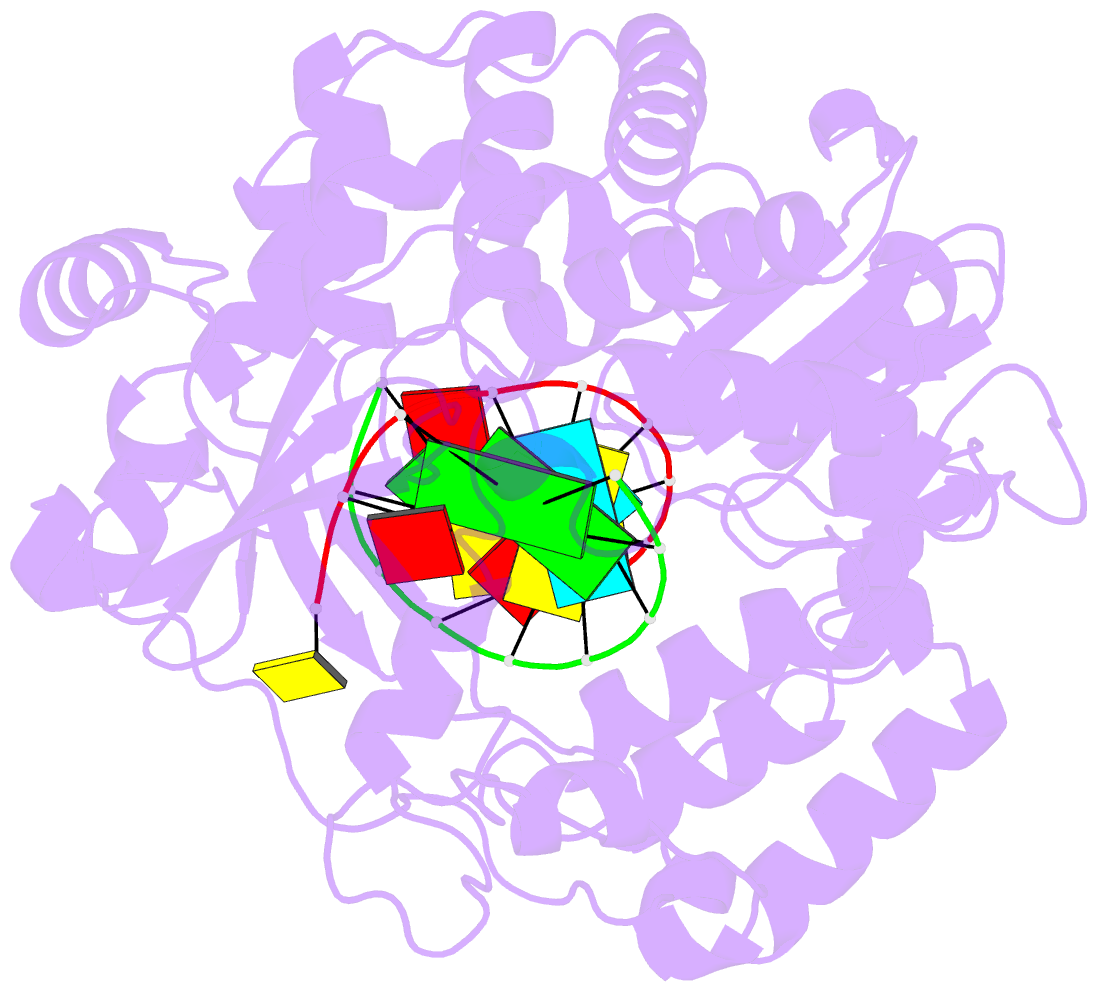
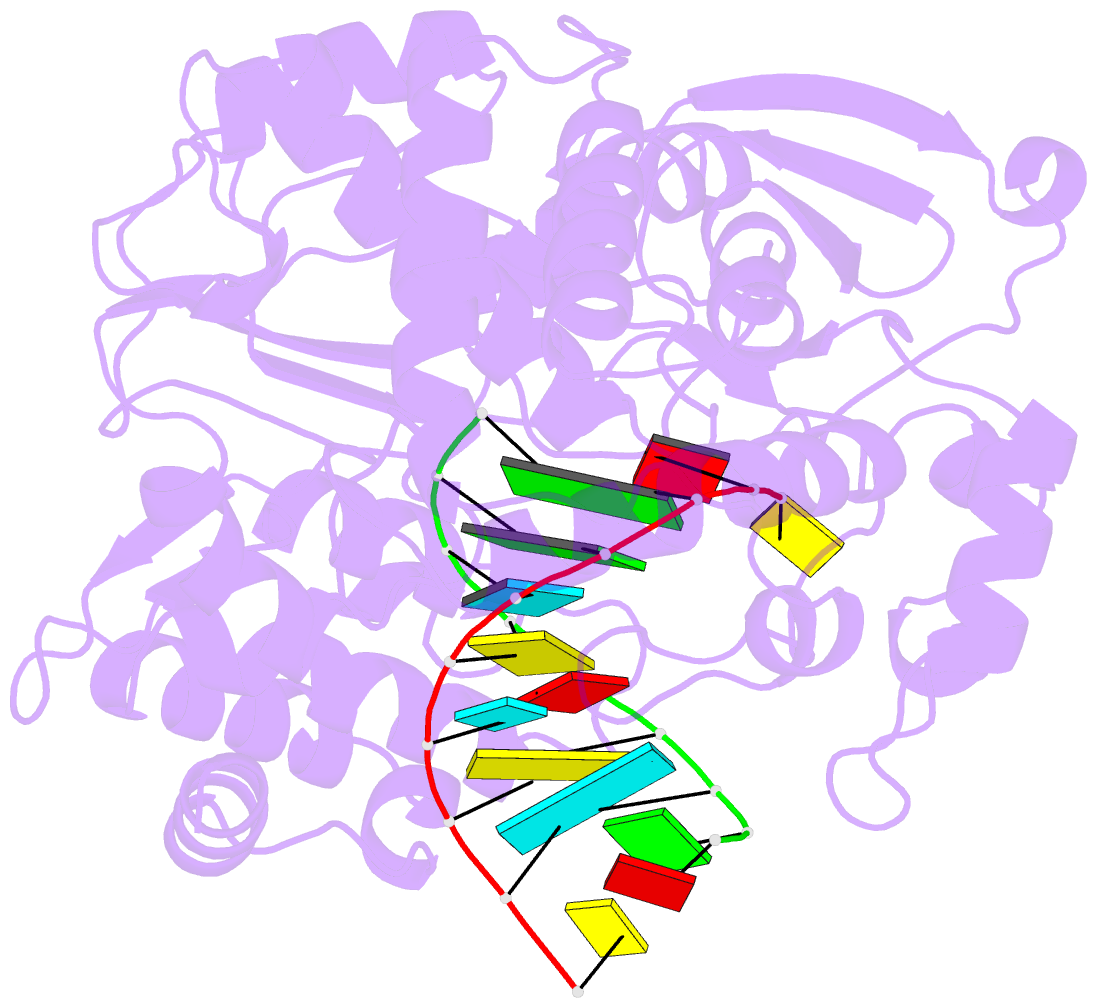
Summary information and primary citation
- PDB-id
- 6lsf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.152 Å)
- Summary
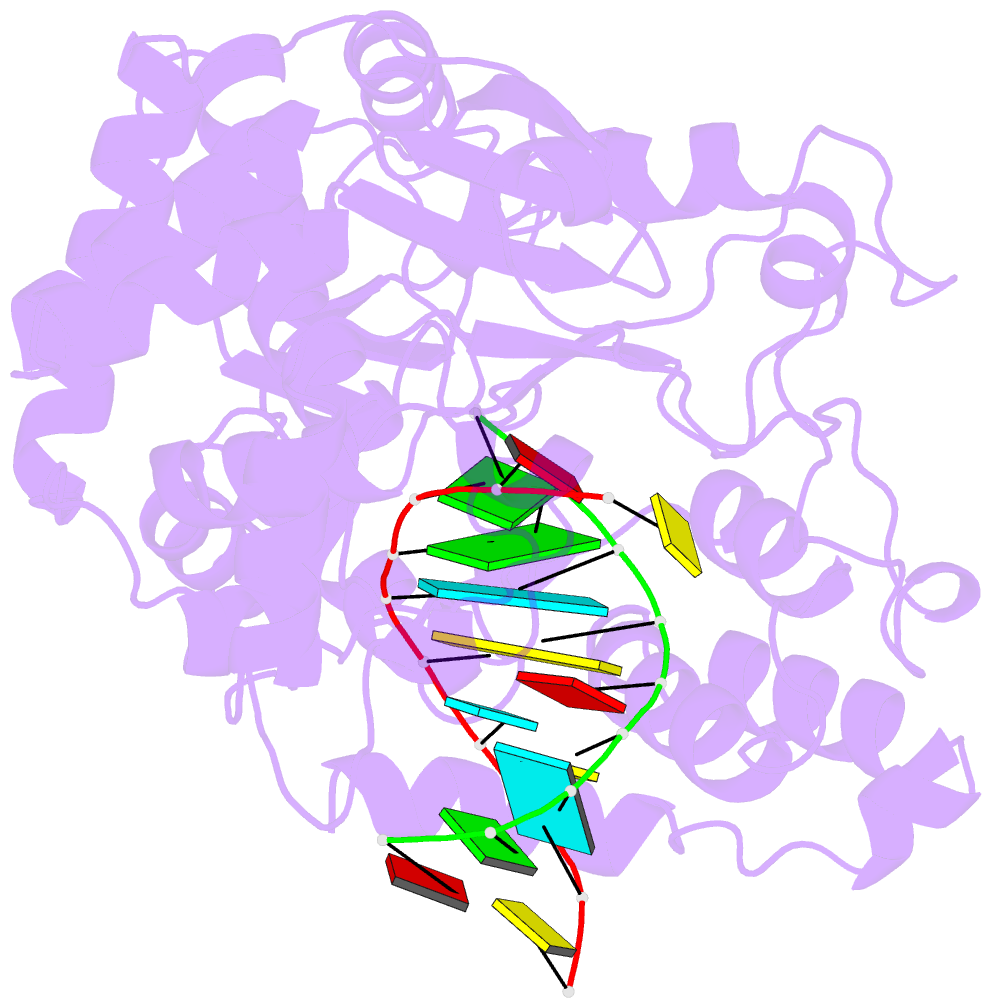
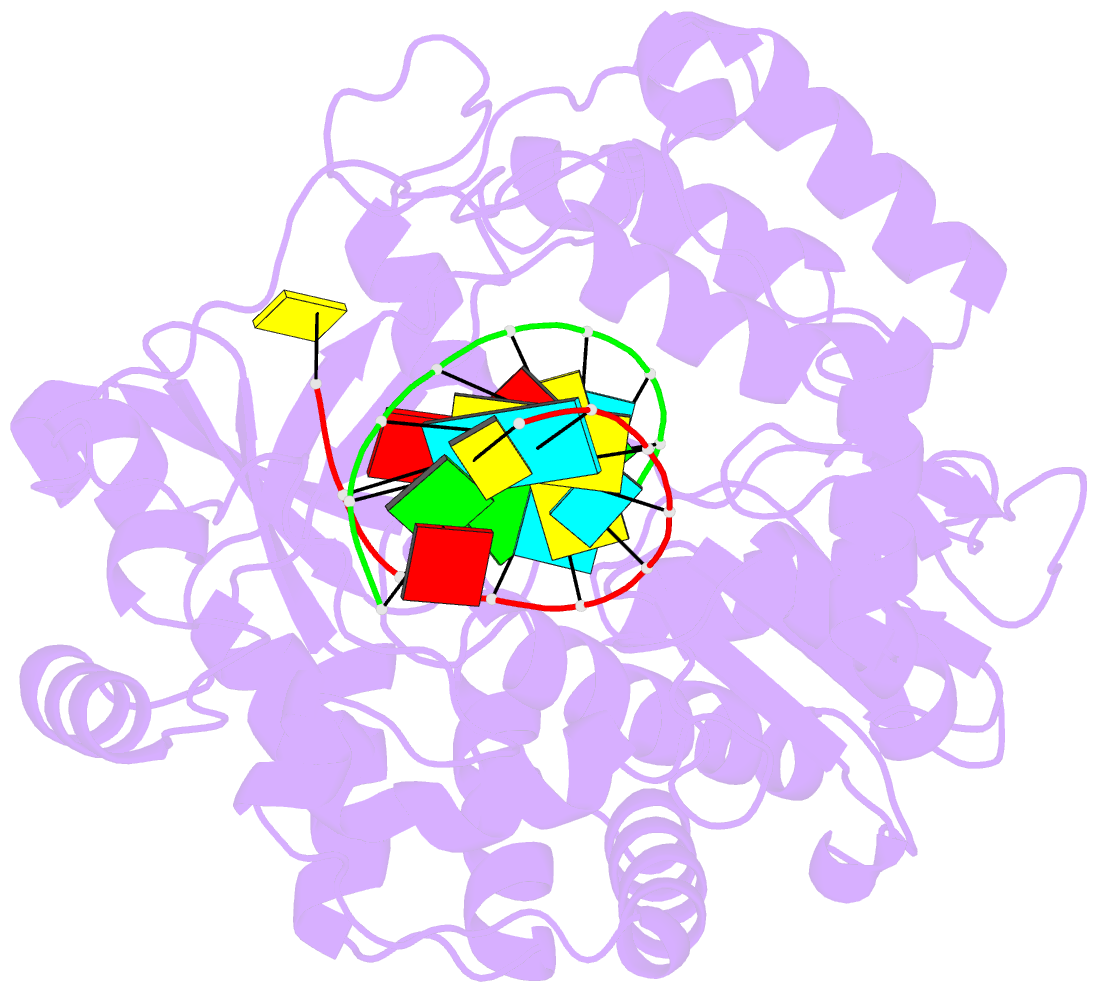
- Crystal structure of the enterovirus 71 polymerase elongation complex (c2s6ra-c2s6rb form)
- Reference
- Wang M, Li R, Shu B, Jing X, Ye HQ, Gong P (2020): "Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures." Nat Commun, 11, 2605. doi: 10.1038/s41467-020-16234-4.
- Abstract
- Each polymerase nucleotide addition cycle is associated with two primary conformational changes of the catalytic complex: the pre-chemistry active site closure and post-chemistry translocation. While active site closure is well interpreted by numerous crystallographic snapshots, translocation intermediates are rarely captured. Here we report three types of intermediate structures in an RNA-dependent RNA polymerase (RdRP). The first two types, captured in forward and reverse translocation events, both highlight the role of RdRP-unique motif G in restricting the RNA template movement, corresponding to the rate-limiting step in translocation. By mutating two critical residues in motif G, we obtain the third type of intermediates that may mimic the transition state of this rate-limiting step, demonstrating a previously unidentified movement of the template strand. We propose that a similar strategy may be utilized by other classes of nucleic acid polymerases to ensure templating nucleotide positioning for efficient catalysis through restricting interactions with template RNA.