Summary information and primary citation
- PDB-id
- 6m6r; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (1.89 Å)
- Summary
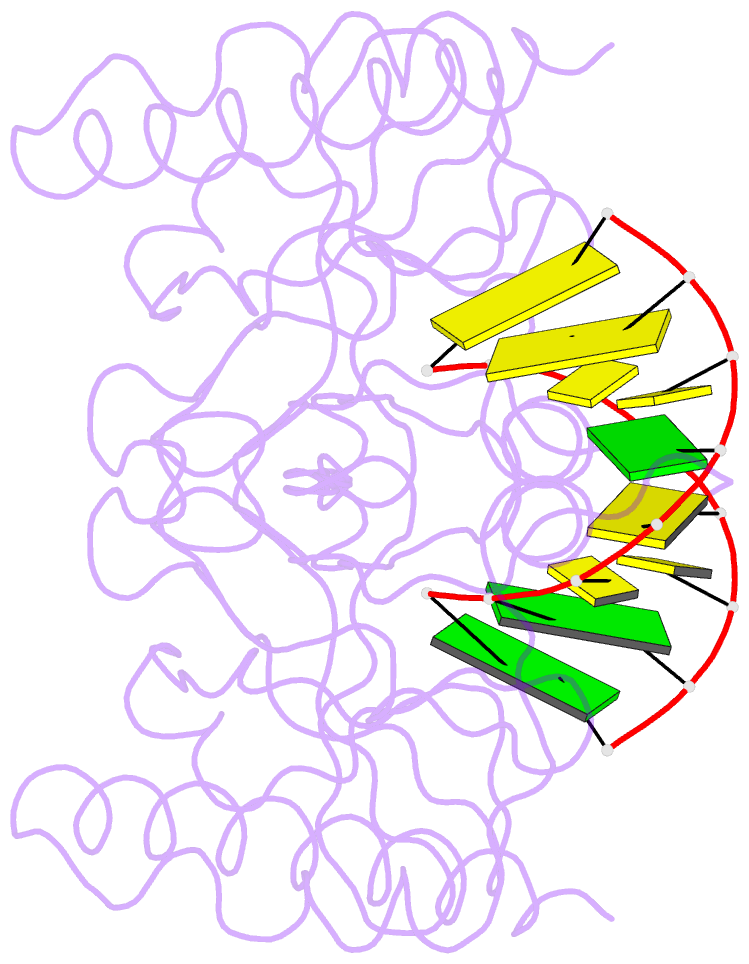
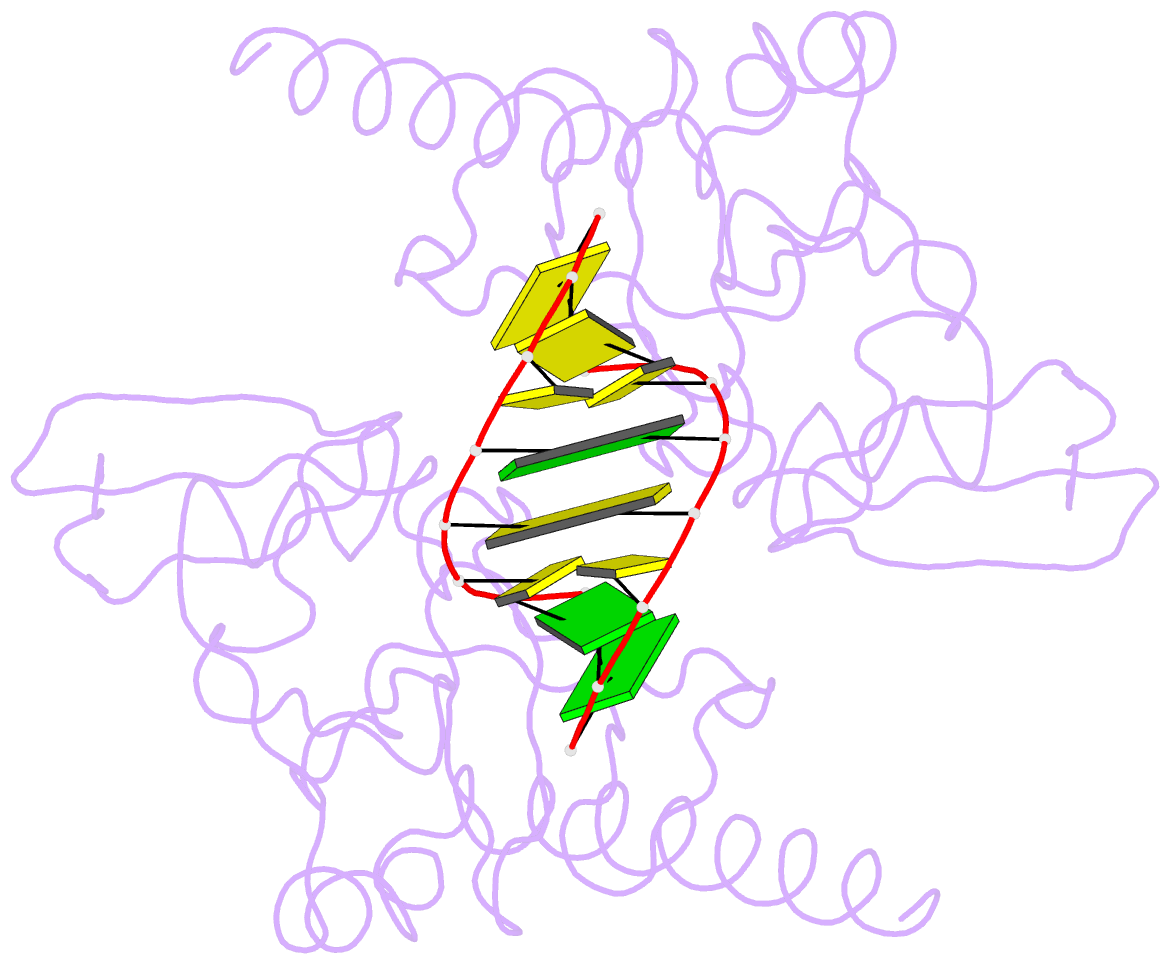
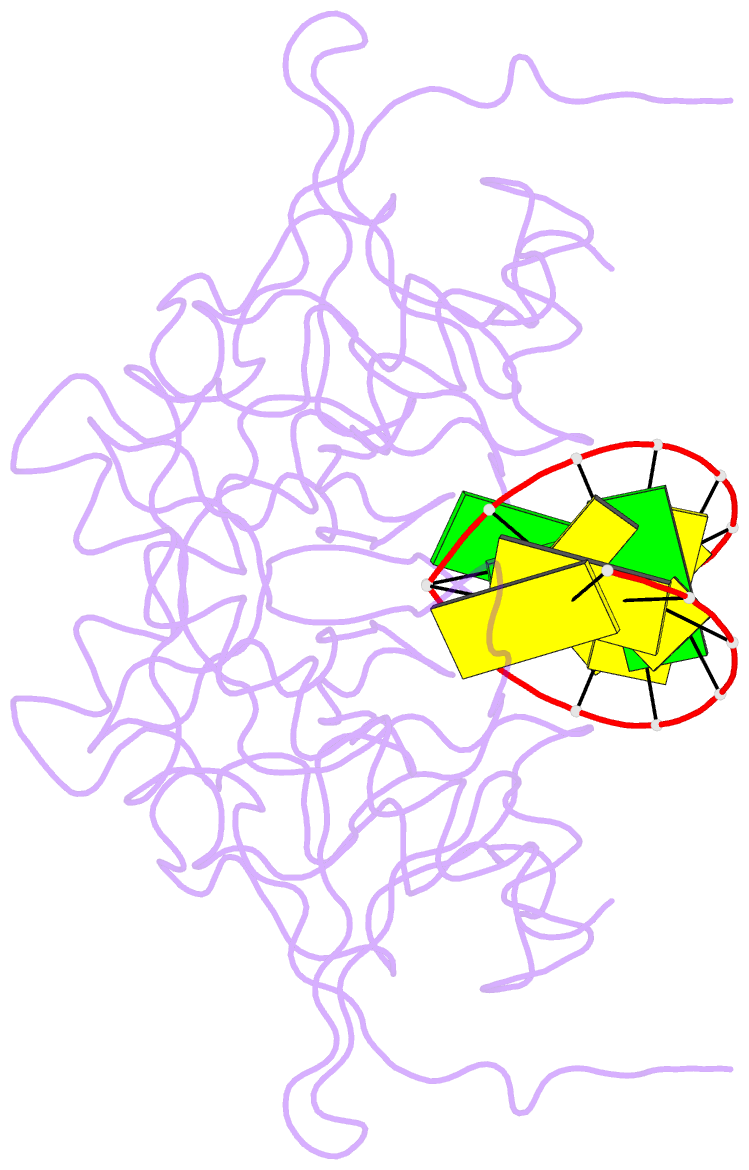
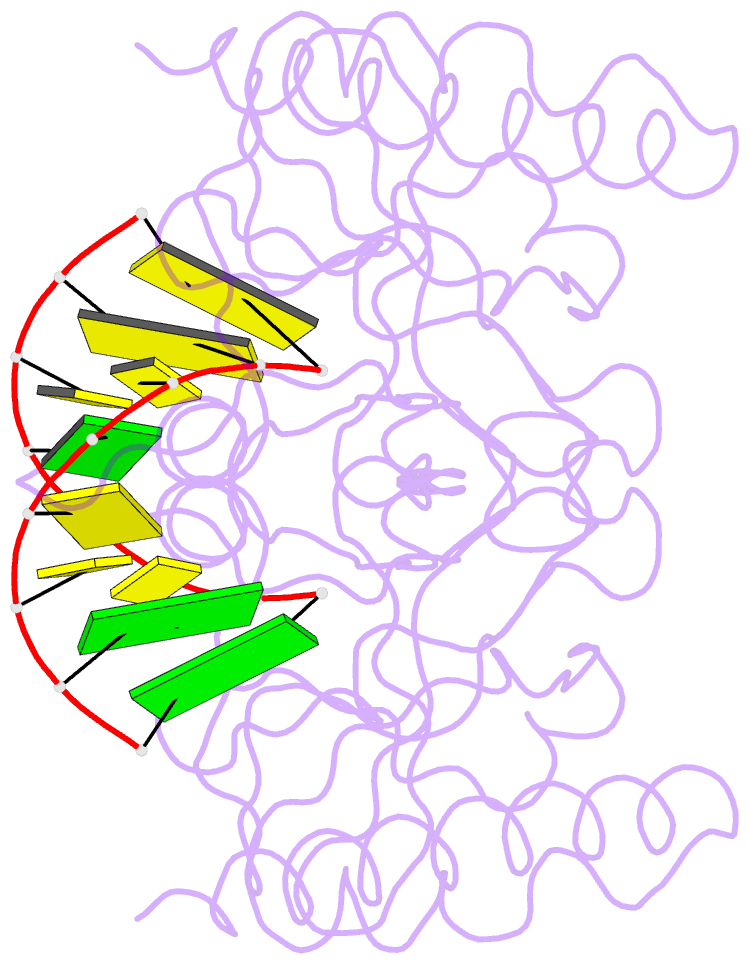
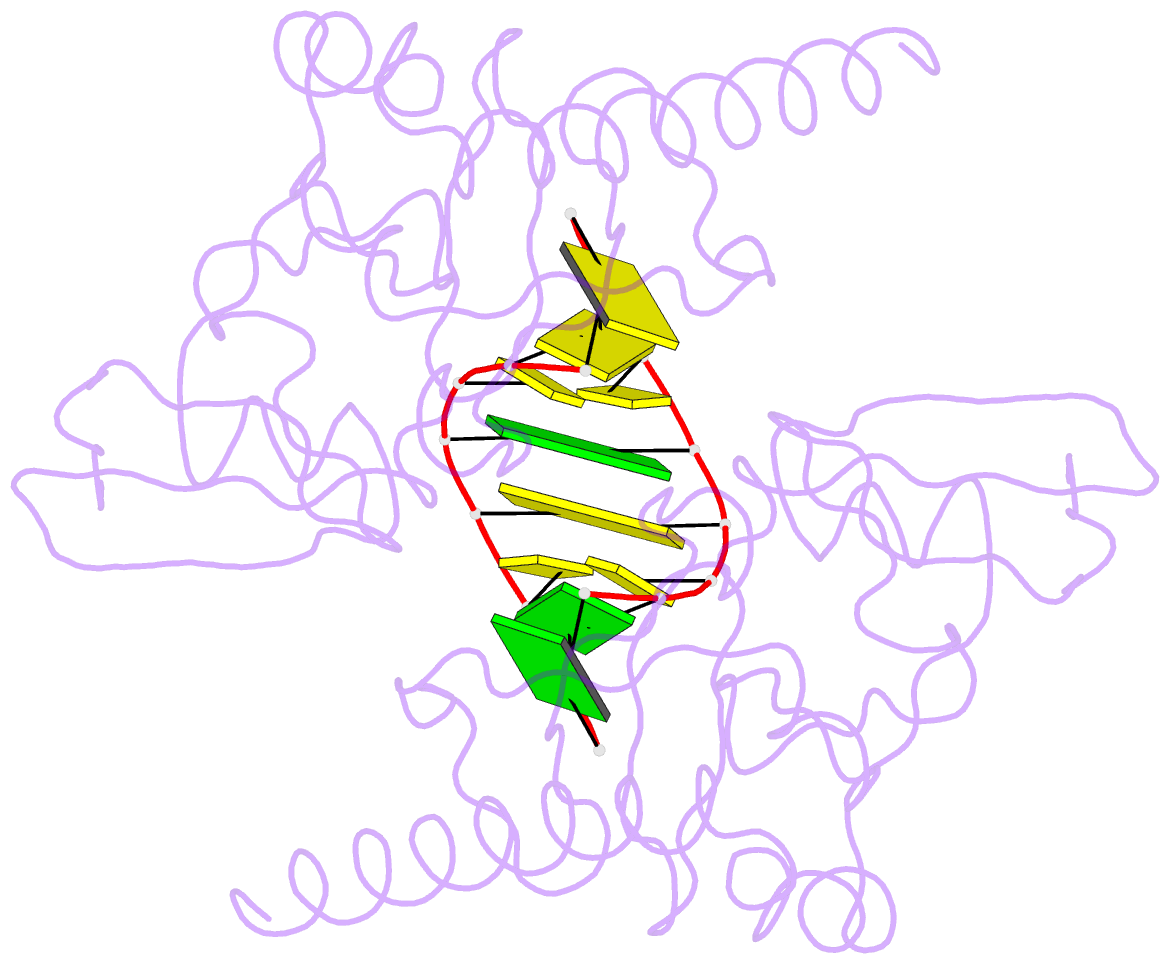
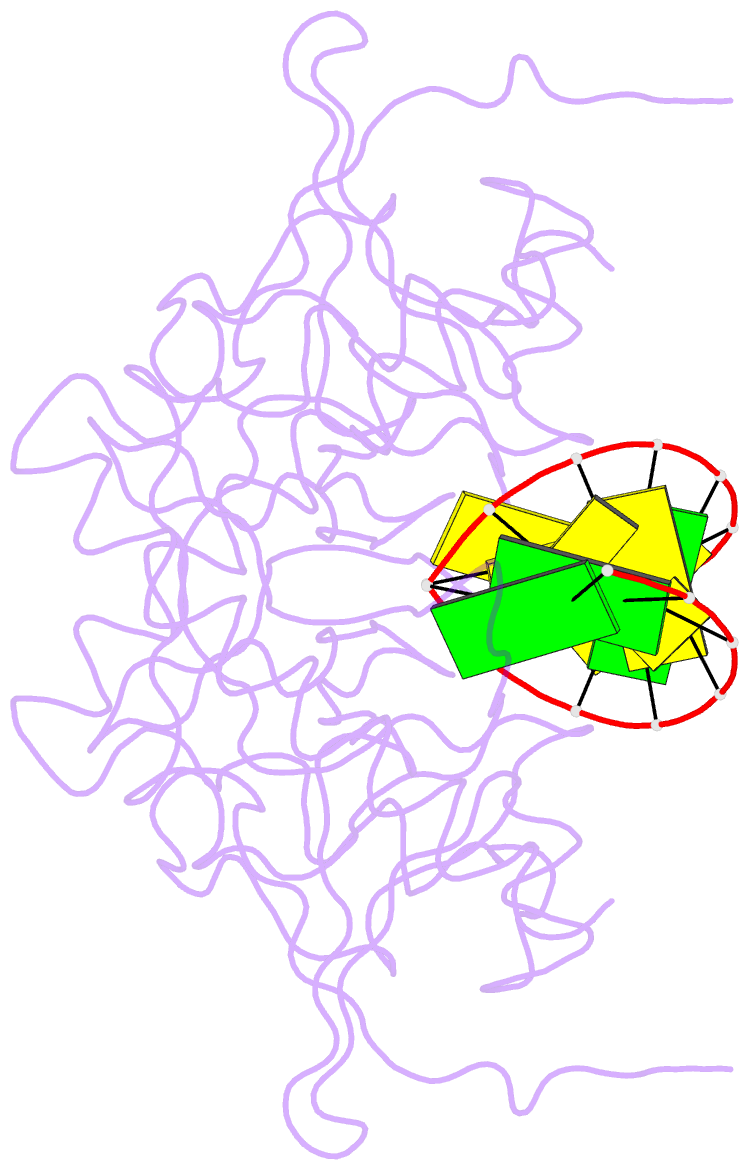
- Crystal structure of caenorhabditis elegans dicer-related helicase 3 (drh-3) c-terminal domain with 5'-ppp 8-mer ssrna
- Reference
- Li K, Zheng J, Wirawan M, Trinh NM, Fedorova O, Griffin PR, Pyle AM, Luo D (2021): "Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3)." Nucleic Acids Res., 49, 9978-9991. doi: 10.1093/nar/gkab712.
- Abstract
- DRH-3 is critically involved in germline development and RNA interference (RNAi) facilitated chromosome segregation via the 22G-siRNA pathway in Caenorhabditis elegans. DRH-3 has similar domain architecture to RIG-I-like receptors (RLRs) and belongs to the RIG-I-like RNA helicase family. The molecular understanding of DRH-3 and its function in endogenous RNAi pathways remains elusive. In this study, we solved the crystal structures of the DRH-3 N-terminal domain (NTD) and the C-terminal domains (CTDs) in complex with 5'-triphosphorylated RNAs. The NTD of DRH-3 adopts a distinct fold of tandem caspase activation and recruitment domains (CARDs) structurally similar to the CARDs of RIG-I and MDA5, suggesting a signaling function in the endogenous RNAi biogenesis. The CTD preferentially recognizes 5'-triphosphorylated double-stranded RNAs bearing the typical features of secondary siRNA transcripts. The full-length DRH-3 displays unique structural dynamics upon binding to RNA duplexes that differ from RIG-I or MDA5. These features of DRH-3 showcase the evolutionary divergence of the Dicer and RLR family of helicases.