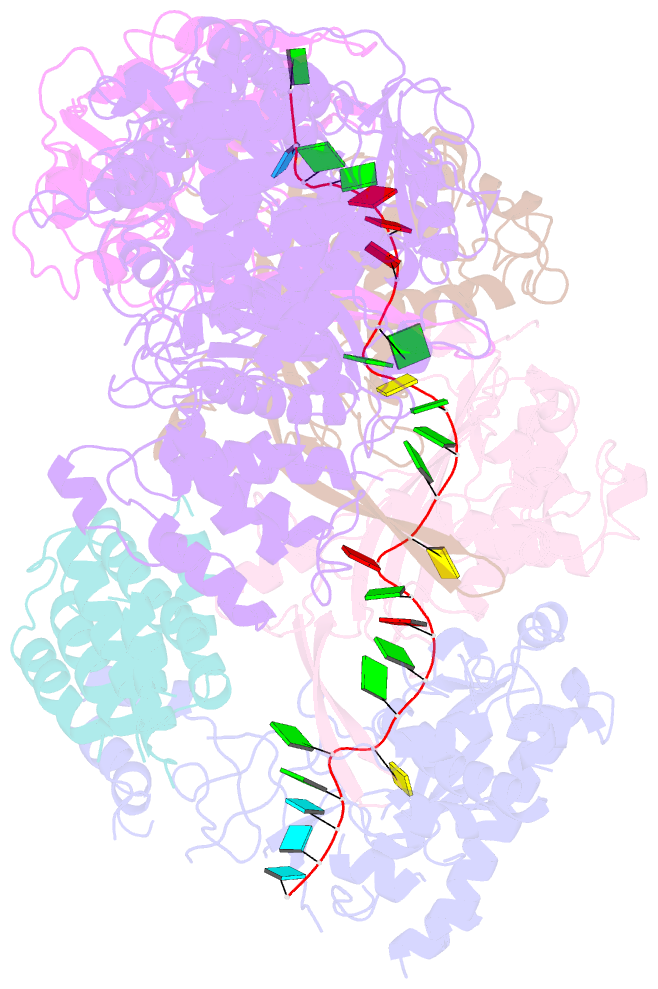
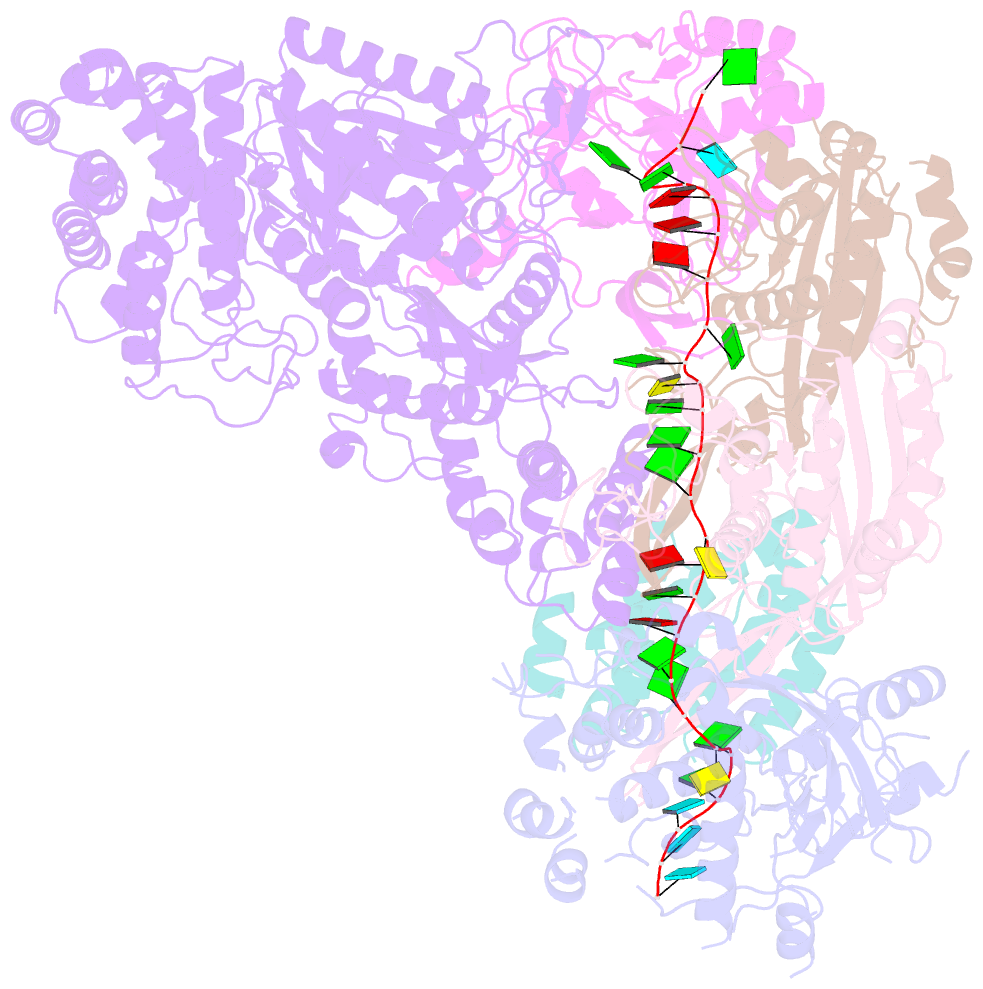
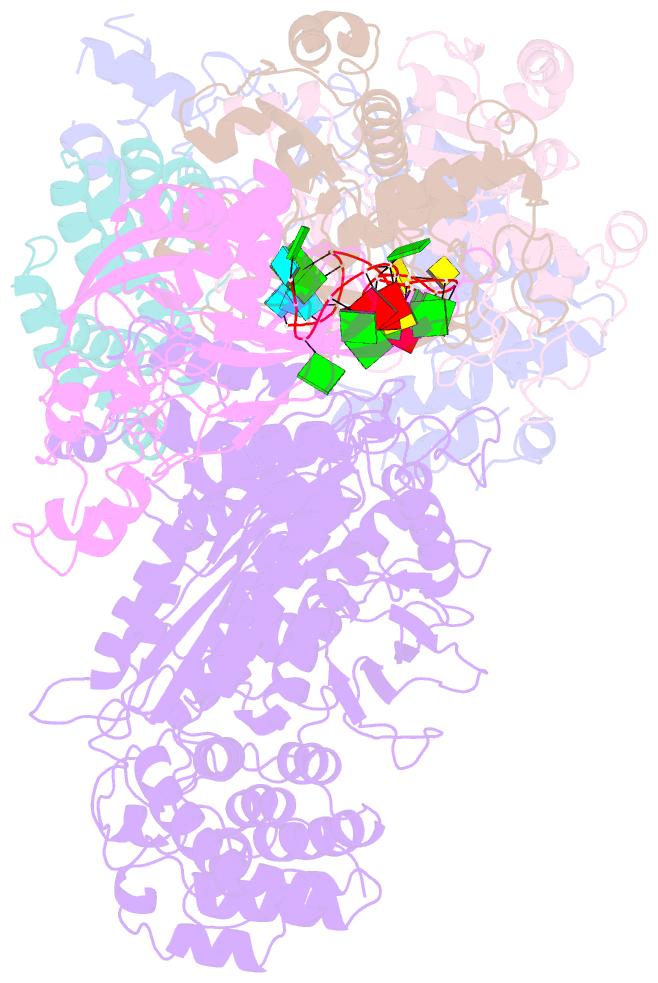
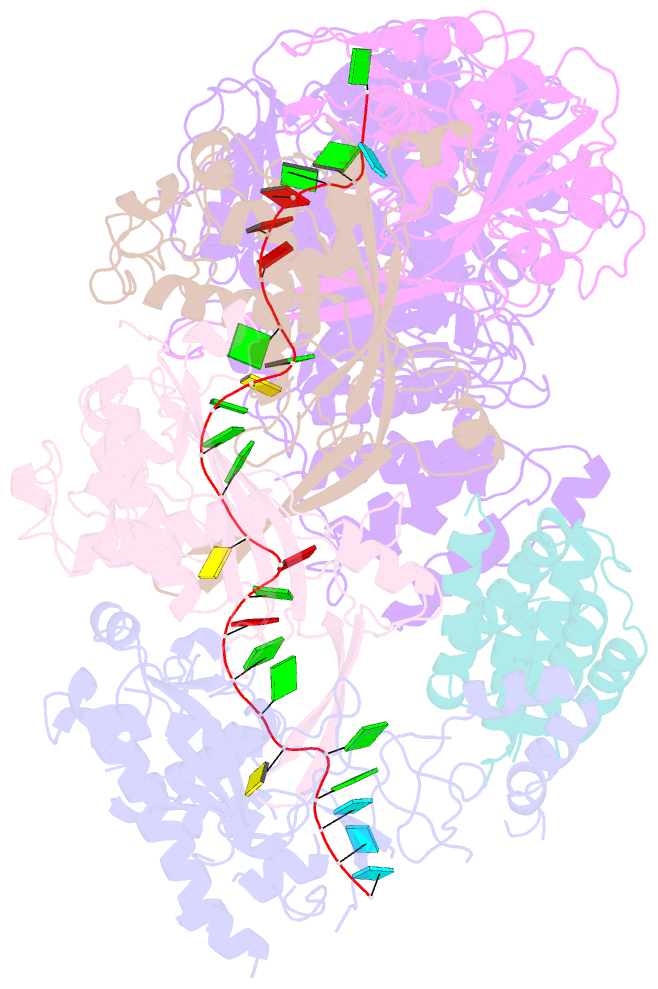
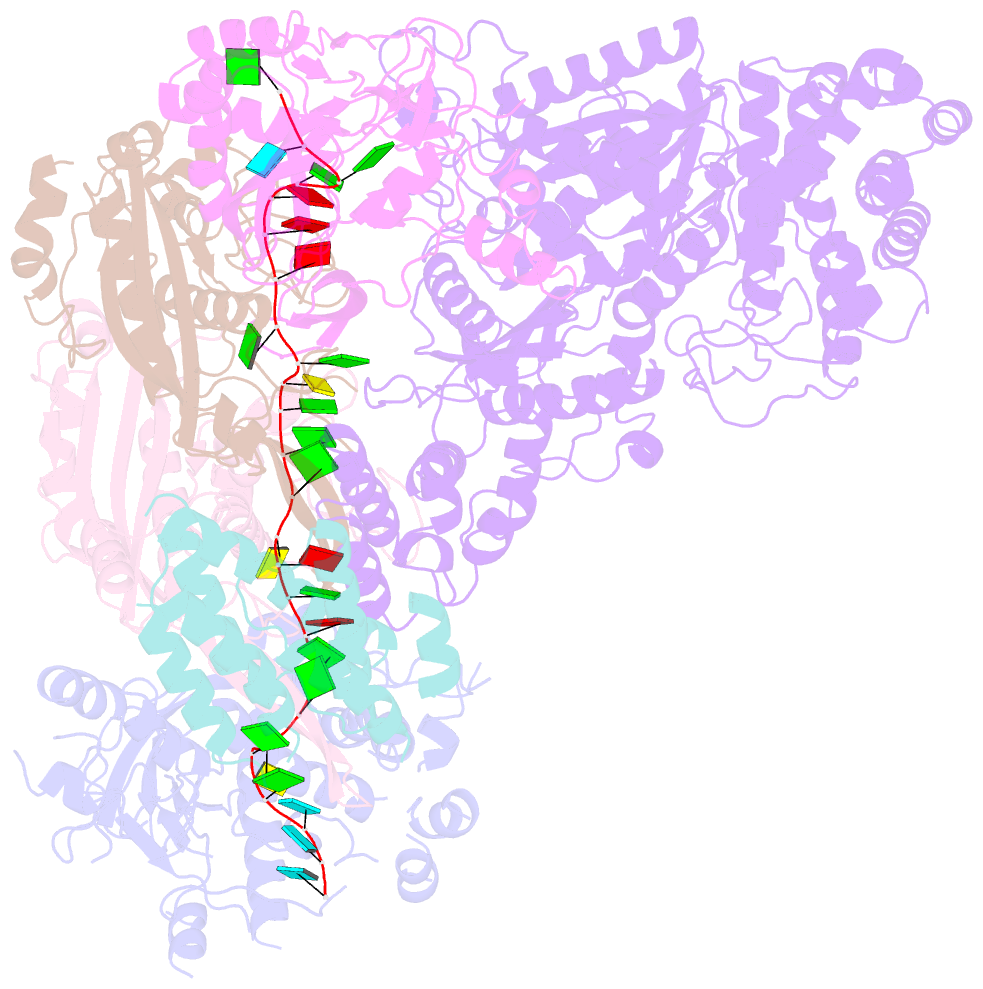
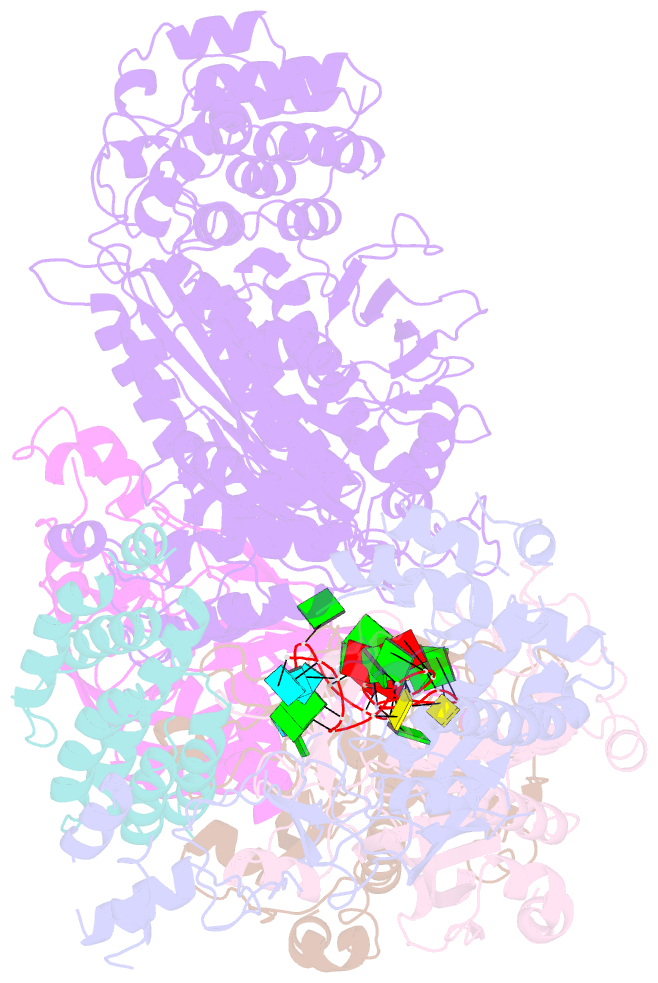
Summary information and primary citation
- PDB-id
- 6muu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.0 Å)
- Summary
- cryo-EM structure of csm-crrna binary complex in type iii-a crispr-cas system
- Reference
- Jia N, Mo CY, Wang C, Eng ET, Marraffini LA, Patel DJ (2019): "Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity." Mol. Cell, 73, 264. doi: 10.1016/j.molcel.2018.11.007.
- Abstract
- Type ΙΙΙ CRISPR-Cas systems provide robust immunity against foreign RNA and DNA by sequence-specific RNase and target RNA-activated sequence-nonspecific DNase and RNase activities. We report on cryo-EM structures of Thermococcus onnurineus CsmcrRNA binary, CsmcrRNA-target RNA and CsmcrRNA-target RNAanti-tag ternary complexes in the 3.1 Å range. The topological features of the crRNA 5'-repeat tag explains the 5'-ruler mechanism for defining target cleavage sites, with accessibility of positions -2 to -5 within the 5'-repeat serving as sensors for avoidance of autoimmunity. The Csm3 thumb elements introduce periodic kinks in the crRNA-target RNA duplex, facilitating cleavage of the target RNA with 6-nt periodicity. Key Glu residues within a Csm1 loop segment of CsmcrRNA adopt a proposed autoinhibitory conformation suggestive of DNase activity regulation. These structural findings, complemented by mutational studies of key intermolecular contacts, provide insights into CsmcrRNA complex assembly, mechanisms underlying RNA targeting and site-specific periodic cleavage, regulation of DNase cleavage activity, and autoimmunity suppression.