Summary information and primary citation
- PDB-id
- 6mwn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA-immune system
- Method
- X-ray (2.838 Å)
- Summary
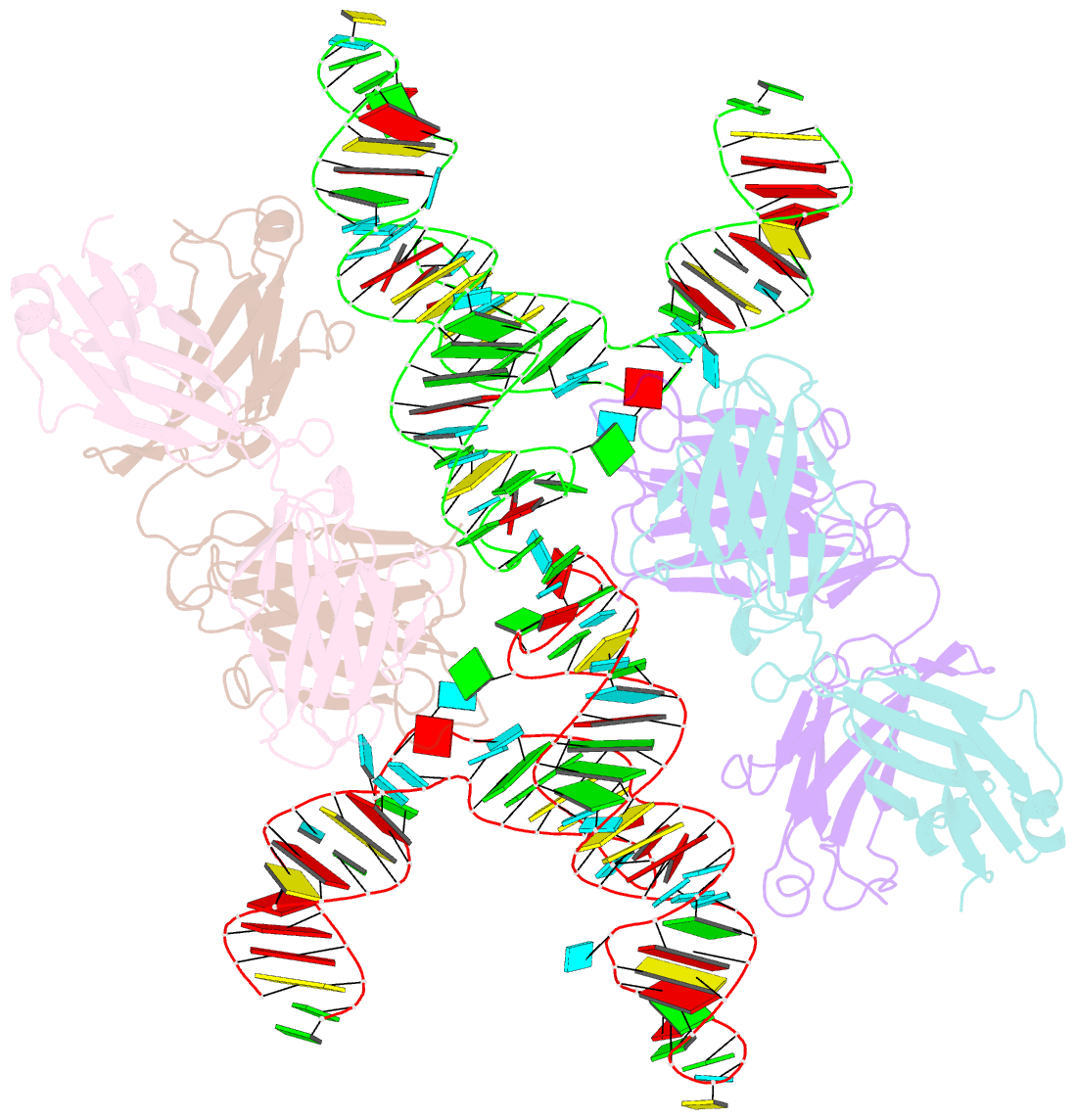
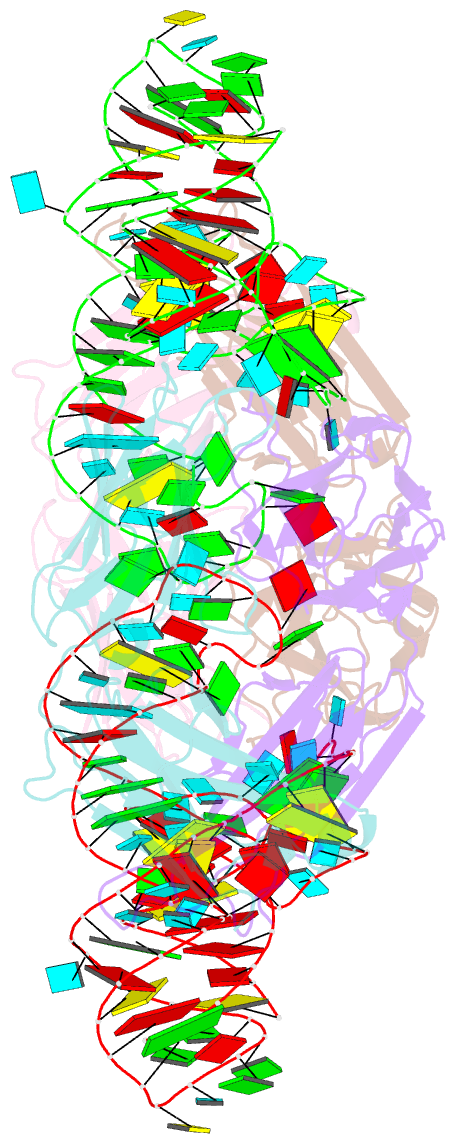
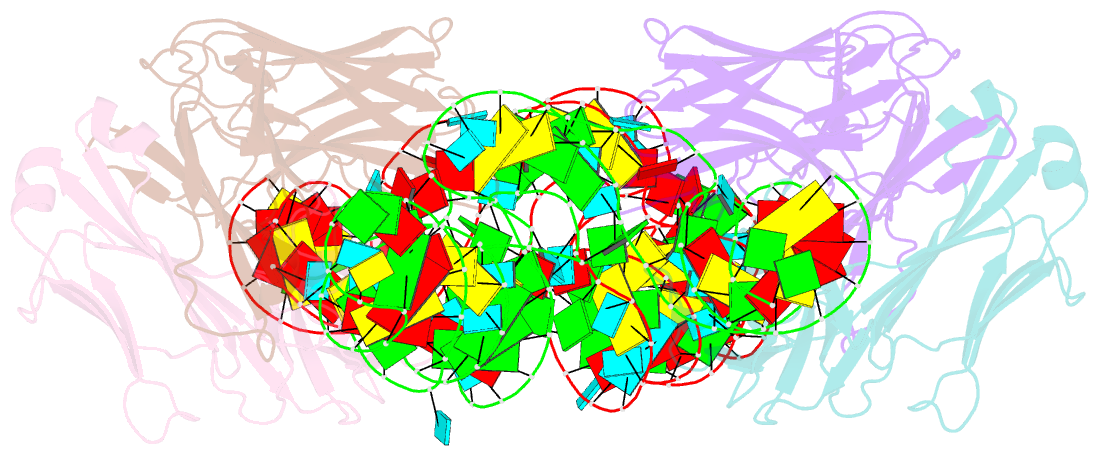
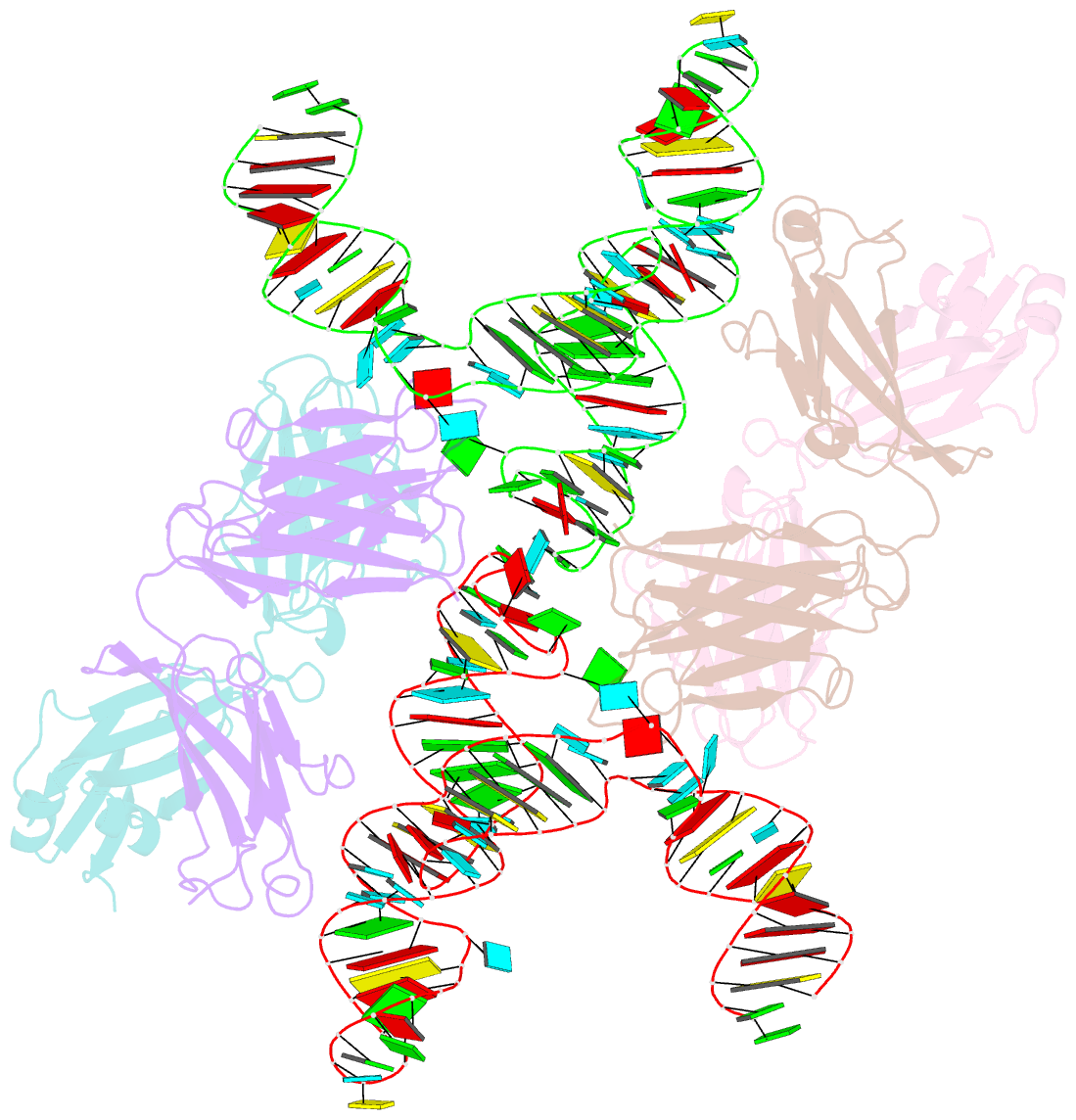
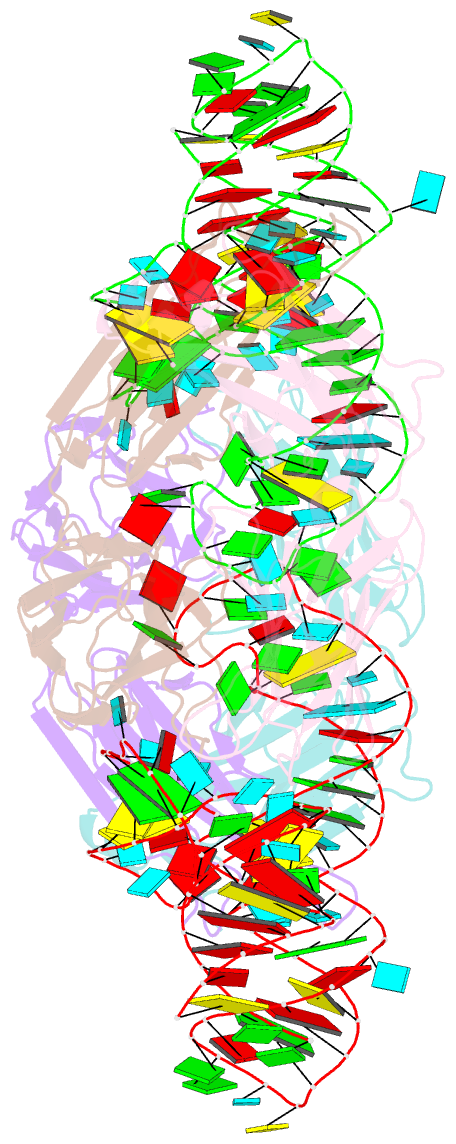
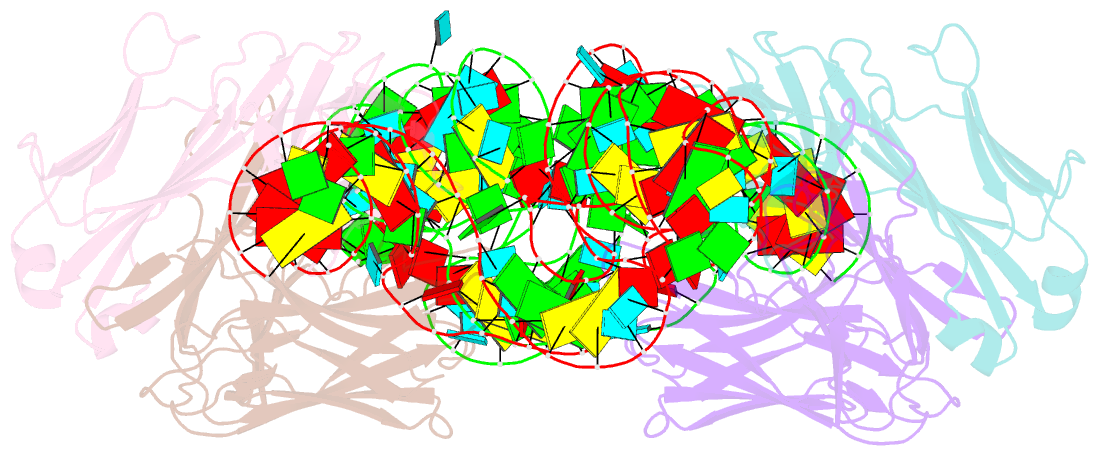
- Crystal structure of hepatitis a virus ires domain v in complex with fab havx
- Reference
- Koirala D, Shao Y, Koldobskaya Y, Fuller JR, Watkins AM, Shelke SA, Pilipenko EV, Das R, Rice PA, Piccirilli JA (2019): "A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites." Nat Commun, 10, 3629. doi: 10.1038/s41467-019-11585-z.
- Abstract
- Picornaviral IRES elements are essential for initiating the cap-independent viral translation. However, three-dimensional structures of these elements remain elusive. Here, we report a 2.84-Å resolution crystal structure of hepatitis A virus IRES domain V (dV) in complex with a synthetic antibody fragment-a crystallization chaperone. The RNA adopts a three-way junction structure, topologically organized by an adenine-rich stem-loop motif. Despite no obvious sequence homology, the dV architecture shows a striking similarity to a circularly permuted form of encephalomyocarditis virus J-K domain, suggesting a conserved strategy for organizing the domain architecture. Recurrence of the motif led us to use homology modeling tools to compute a 3-dimensional structure of the corresponding domain of foot-and-mouth disease virus, revealing an analogous domain organizing motif. The topological conservation observed among these IRESs and other viral domains implicates a structured three-way junction as an architectural scaffold to pre-organize helical domains for recruiting the translation initiation machinery.