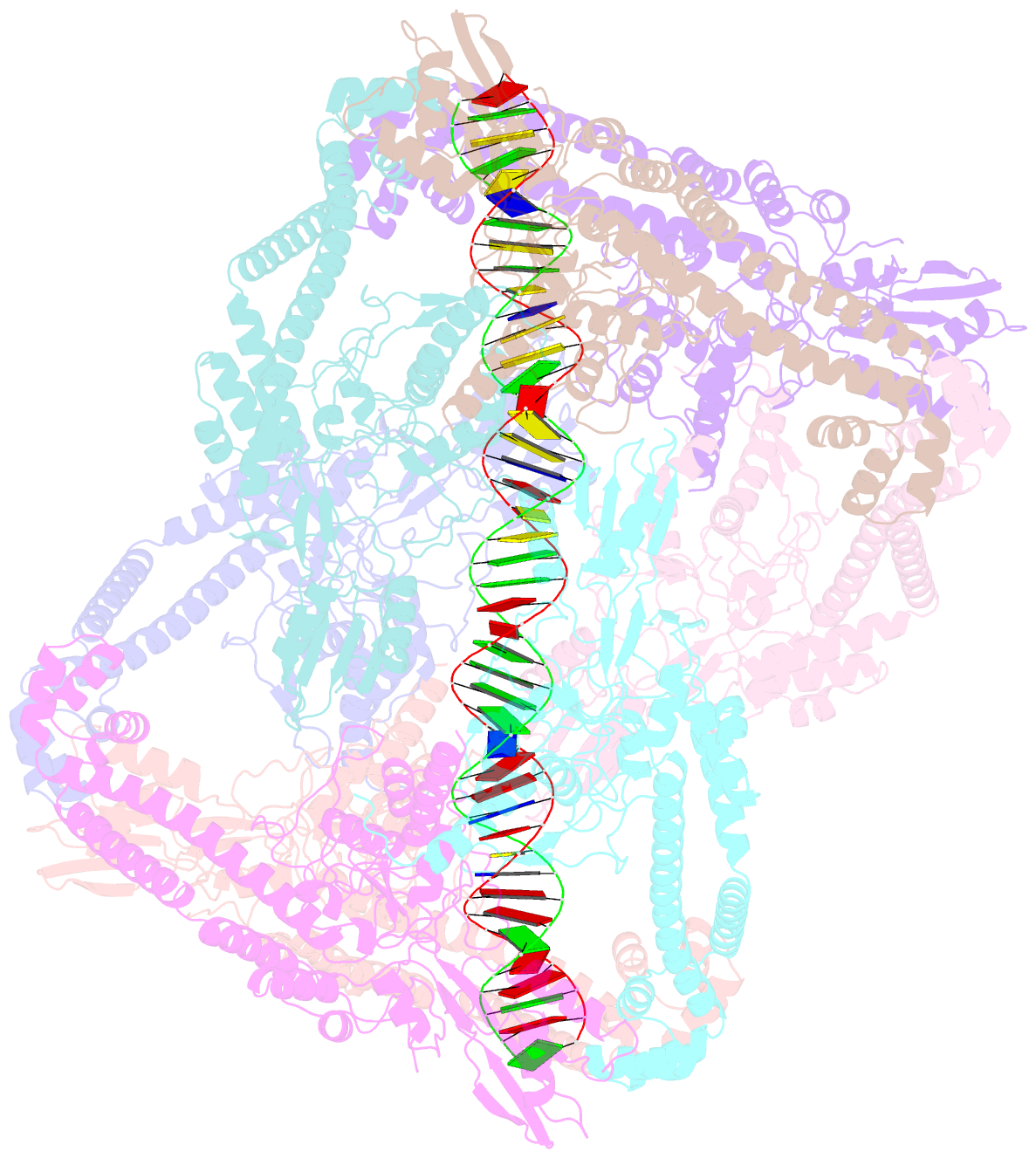
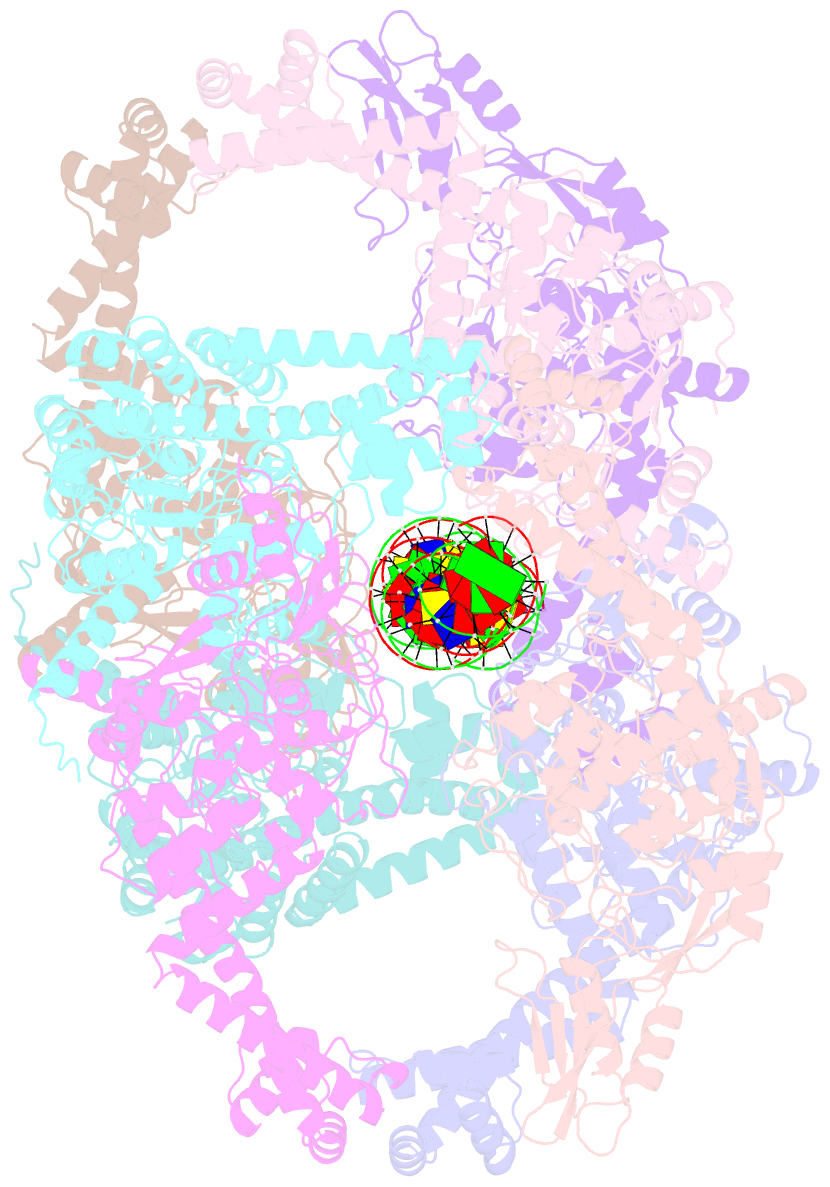
Summary information and primary citation
- PDB-id
- 6n1p; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-DNA
- Method
- cryo-EM (6.35 Å)
- Summary
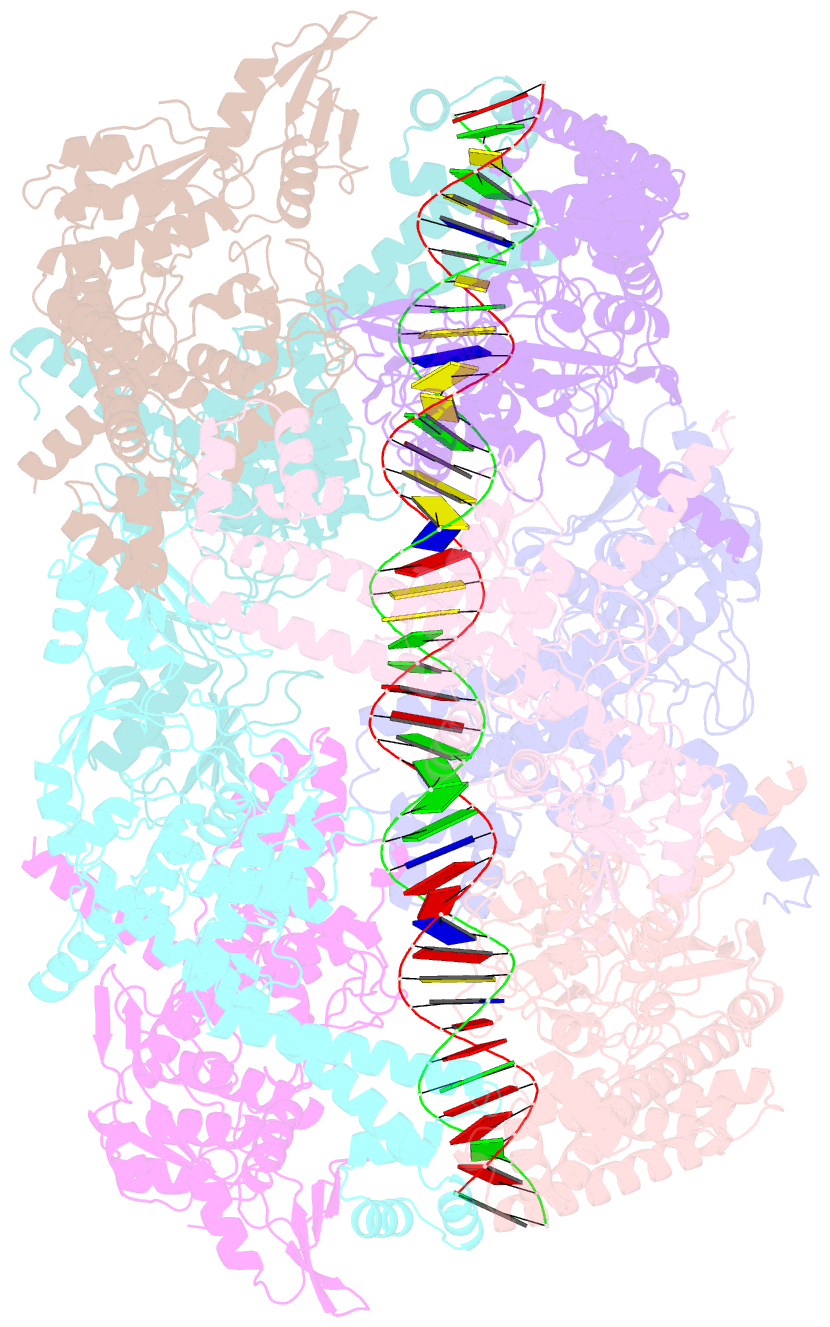
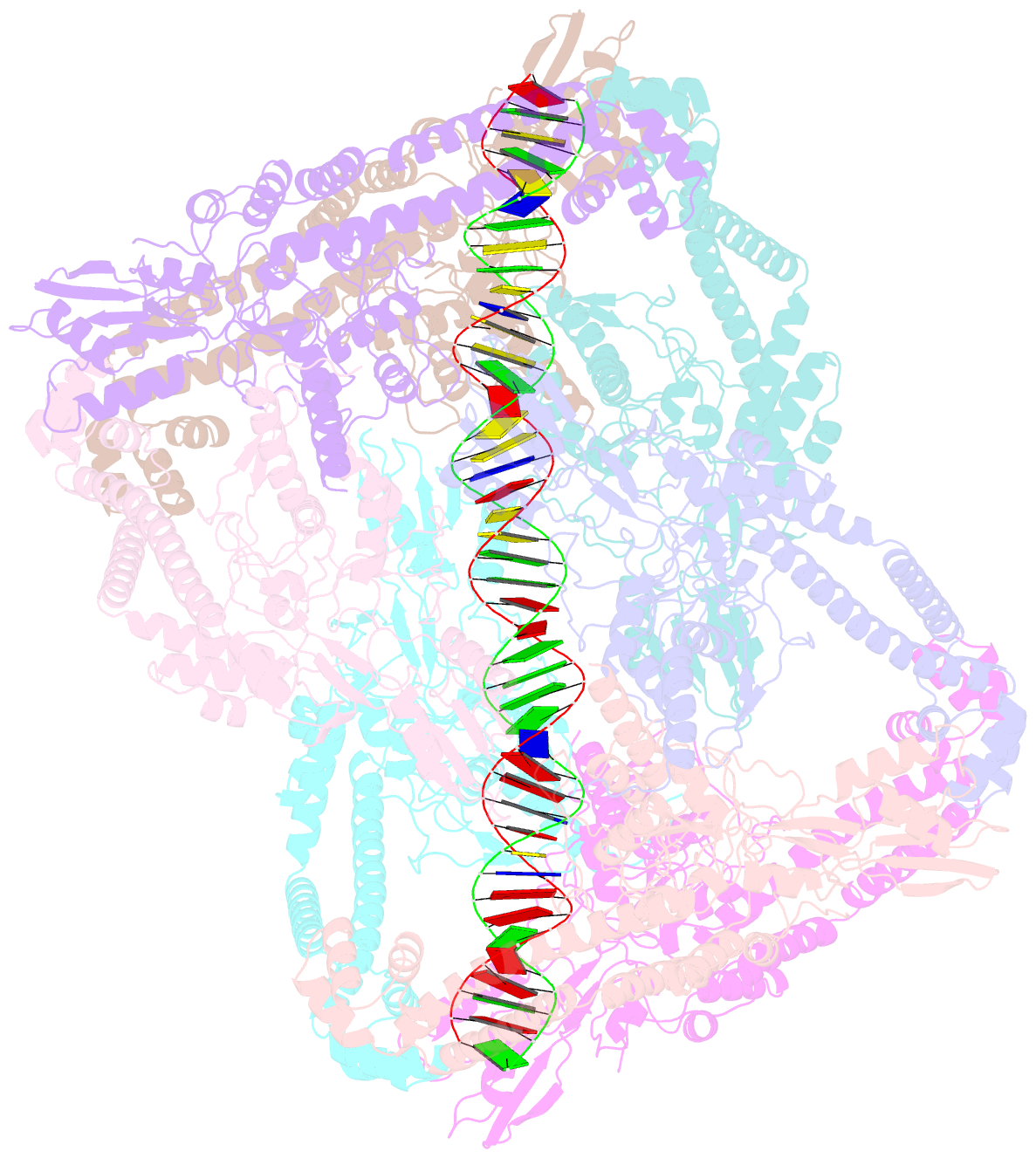
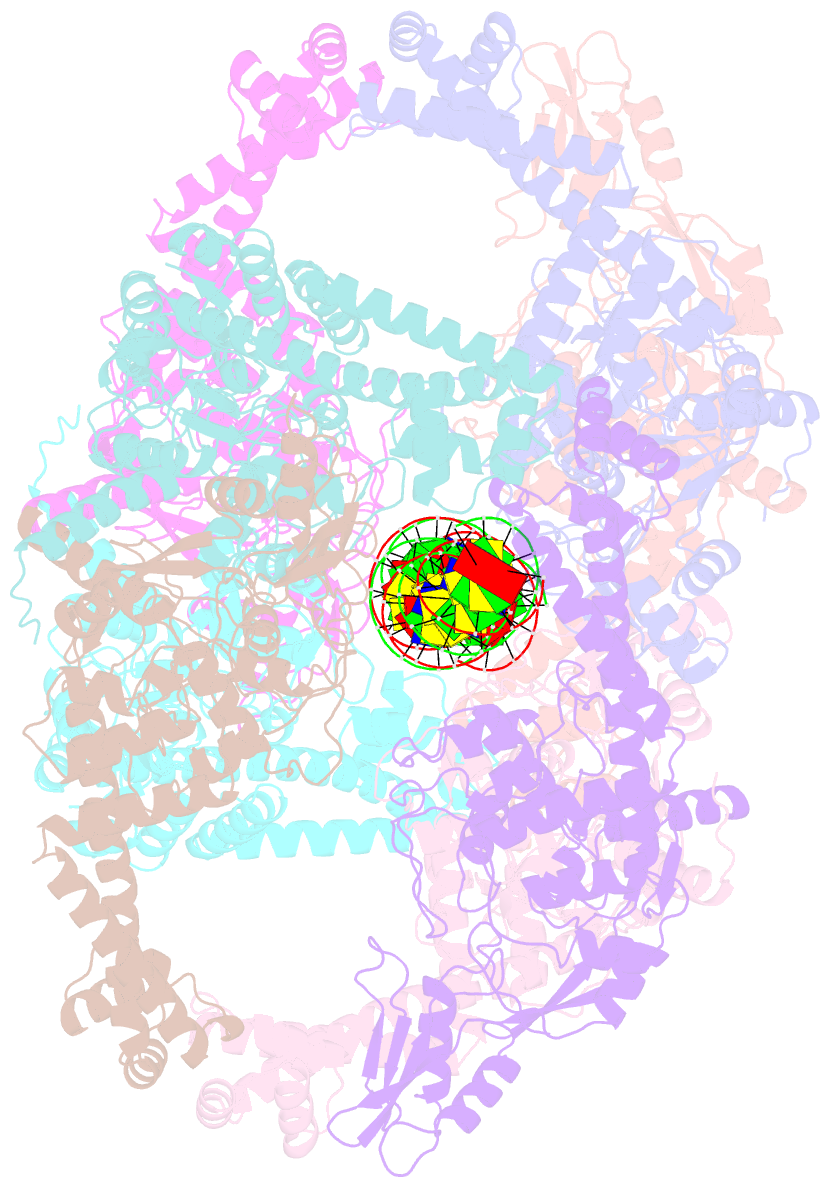
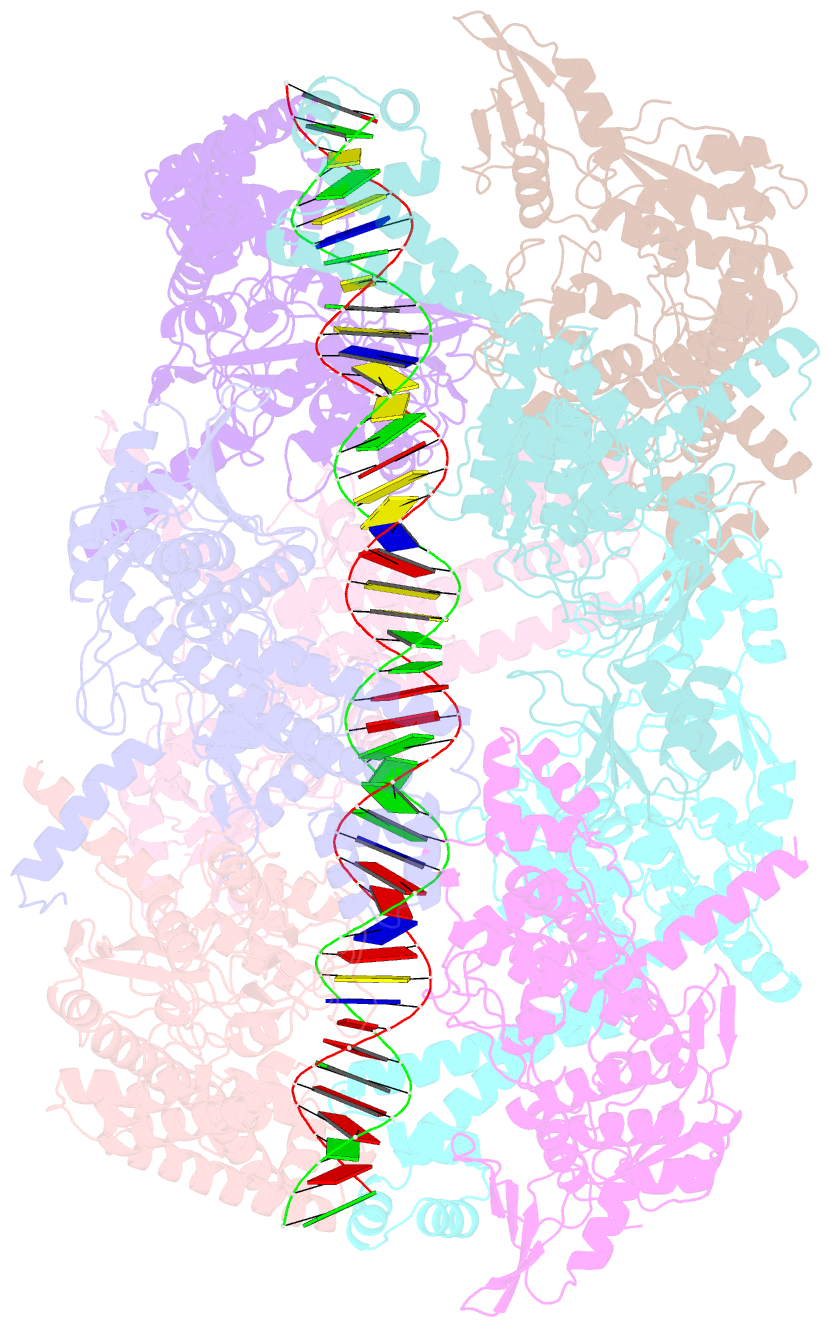
- Dihedral oligomeric complex of gyra n-terminal fragment with DNA, solved by cryoem in c2 symmetry
- Reference
- Soczek KM, Grant T, Rosenthal PB, Mondragon A (2018): "CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage." Elife, 7. doi: 10.7554/eLife.41215.
- Abstract
- Gyrase is a unique type IIA topoisomerase that uses ATP hydrolysis to maintain the negatively supercoiled state of bacterial DNA. In order to perform its function, gyrase undergoes a sequence of conformational changes that consist of concerted gate openings, DNA cleavage, and DNA strand passage events. Structures where the transported DNA molecule (T-segment) is trapped by the A subunit have not been observed. Here we present the cryoEM structures of two oligomeric complexes of open gyrase A dimers and DNA. The protein subunits in these complexes were solved to 4 Å and 5.2 Å resolution. One of the complexes traps a linear DNA molecule, a putative T-segment, which interacts with the open gyrase A dimers in two states, representing steps either prior to or after passage through the DNA-gate. The structures locate the T-segment in important intermediate conformations of the catalytic cycle and provide insights into gyrase-DNA interactions and mechanism.