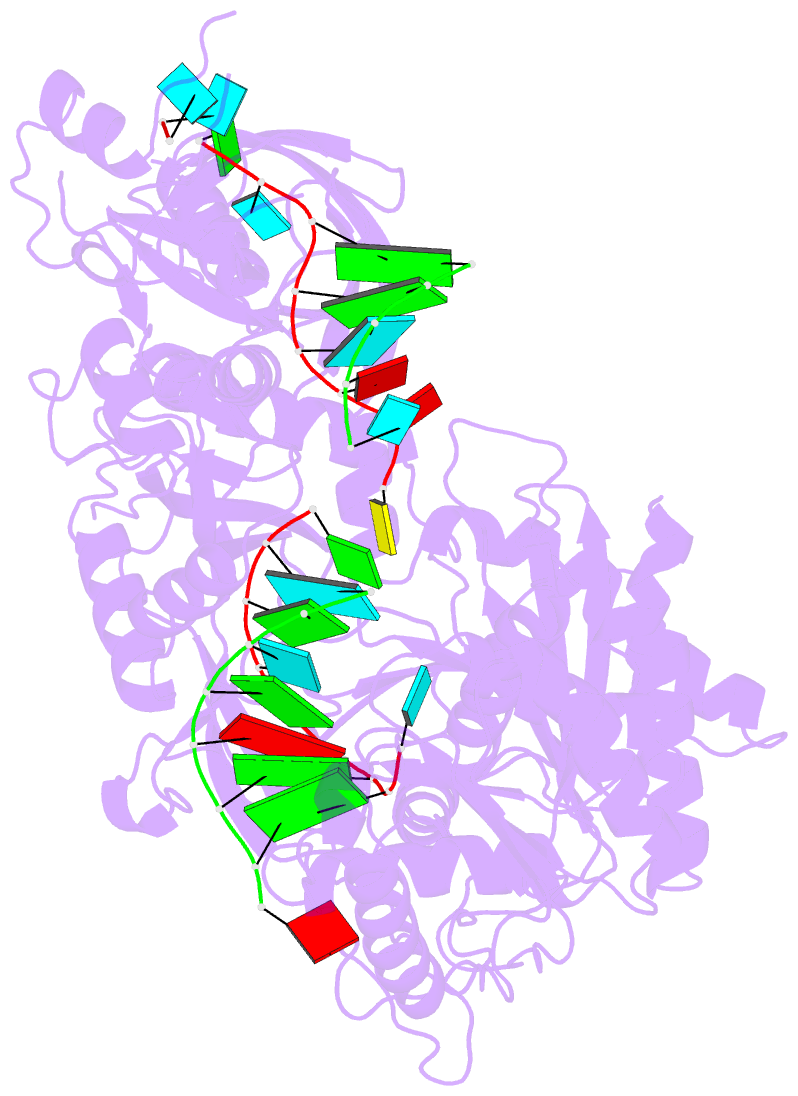
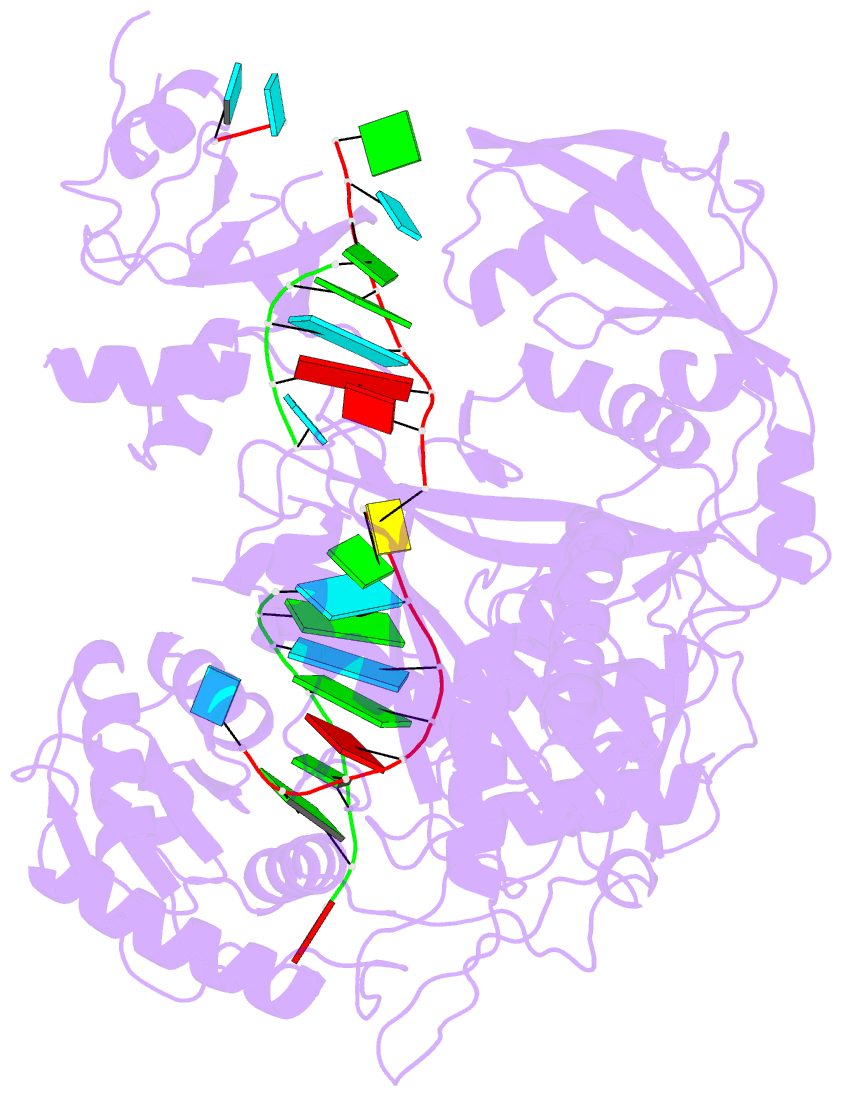
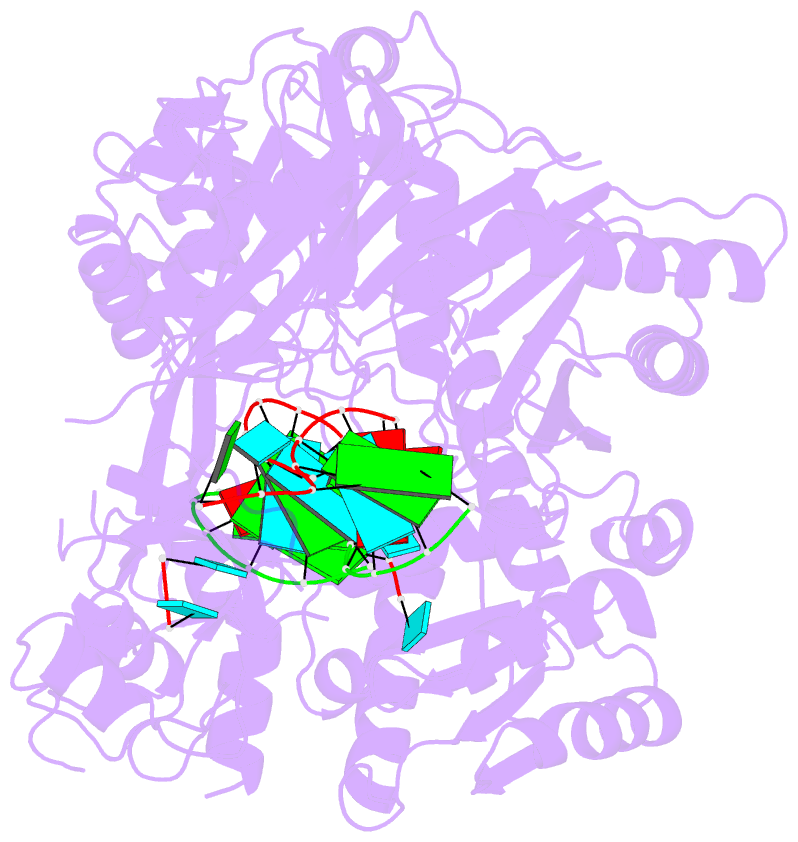
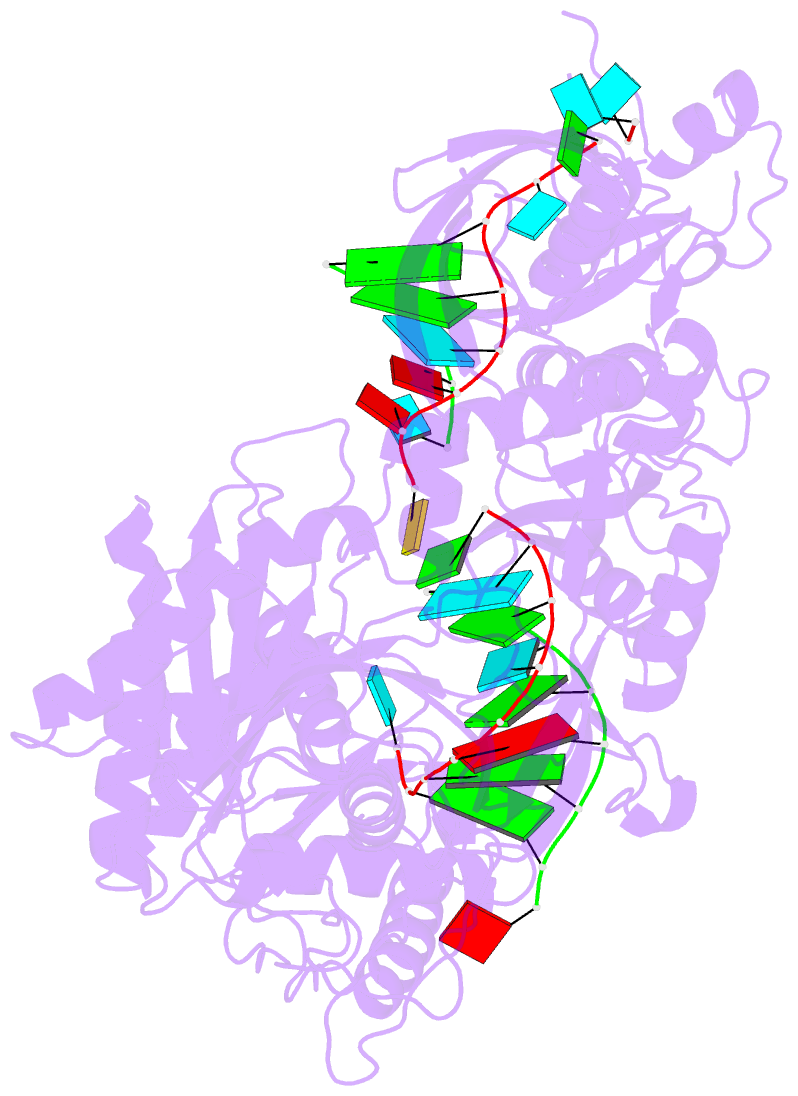
Summary information and primary citation
- PDB-id
- 6n4o; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.899 Å)
- Summary
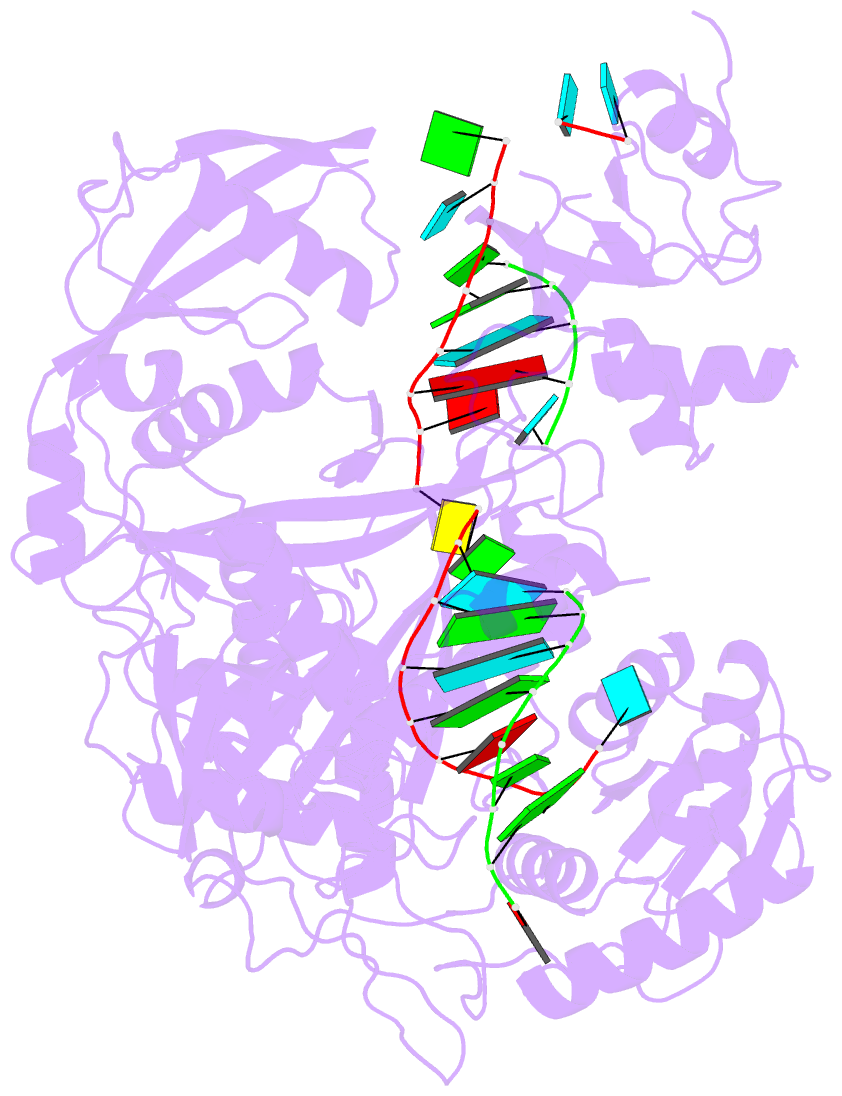
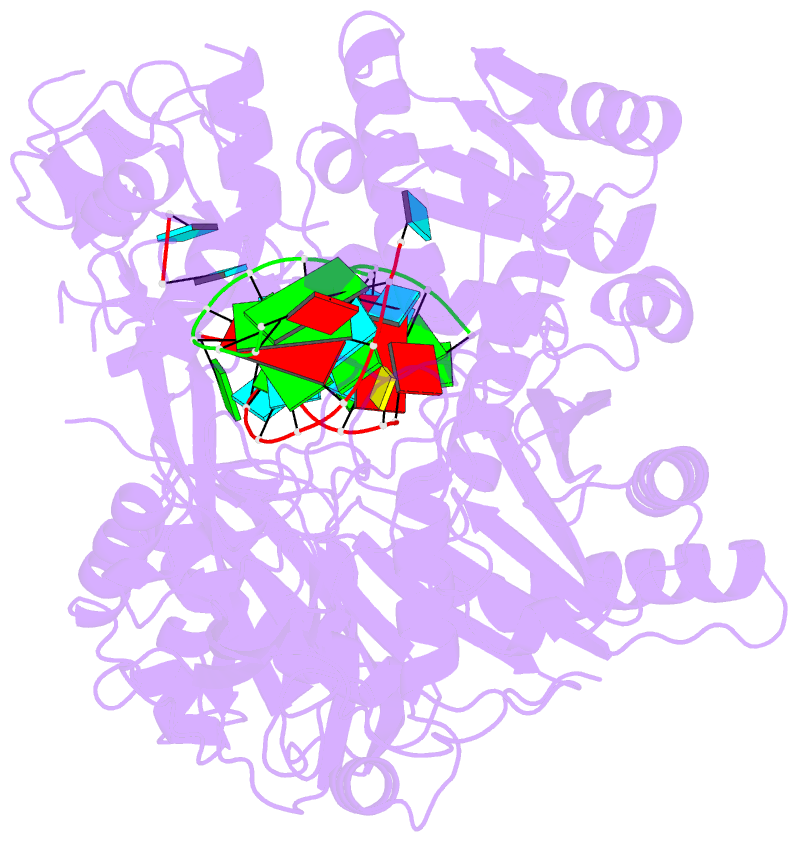
- Human argonaute2-mir-122 bound to a seed and supplementary paired target
- Reference
- Sheu-Gruttadauria J, Xiao Y, Gebert LF, MacRae IJ (2019): "Beyond the seed: structural basis for supplementary microRNA targeting by human Argonaute2." Embo J., 38, e101153. doi: 10.15252/embj.2018101153.
- Abstract
- microRNAs (miRNAs) guide Argonaute proteins to mRNAs targeted for repression. Target recognition occurs primarily through the miRNA seed region, composed of guide (g) nucleotides g2-g8. However, nucleotides beyond the seed are also important for some known miRNA-target interactions. Here, we report the structure of human Argonaute2 (Ago2) engaged with a target RNA recognized through both miRNA seed and supplementary (g13-g16) regions. Ago2 creates a "supplementary chamber" that accommodates up to five miRNA-target base pairs. Seed and supplementary chambers are adjacent to each other and can be bridged by an unstructured target loop of 1-15 nucleotides. Opening of the supplementary chamber may be constrained by tension in the miRNA 3' tail, as increases in miRNA length stabilize supplementary interactions. Contrary to previous reports, we demonstrate that optimal supplementary interactions can increase target affinity > 20-fold. These results provide a mechanism for extended miRNA targeting, suggest a function for 3' isomiRs in tuning miRNA targeting specificity, and indicate that supplementary interactions may contribute more to target recognition than is widely appreciated.