Summary information and primary citation
- PDB-id
- 6n7x; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.6 Å)
- Summary
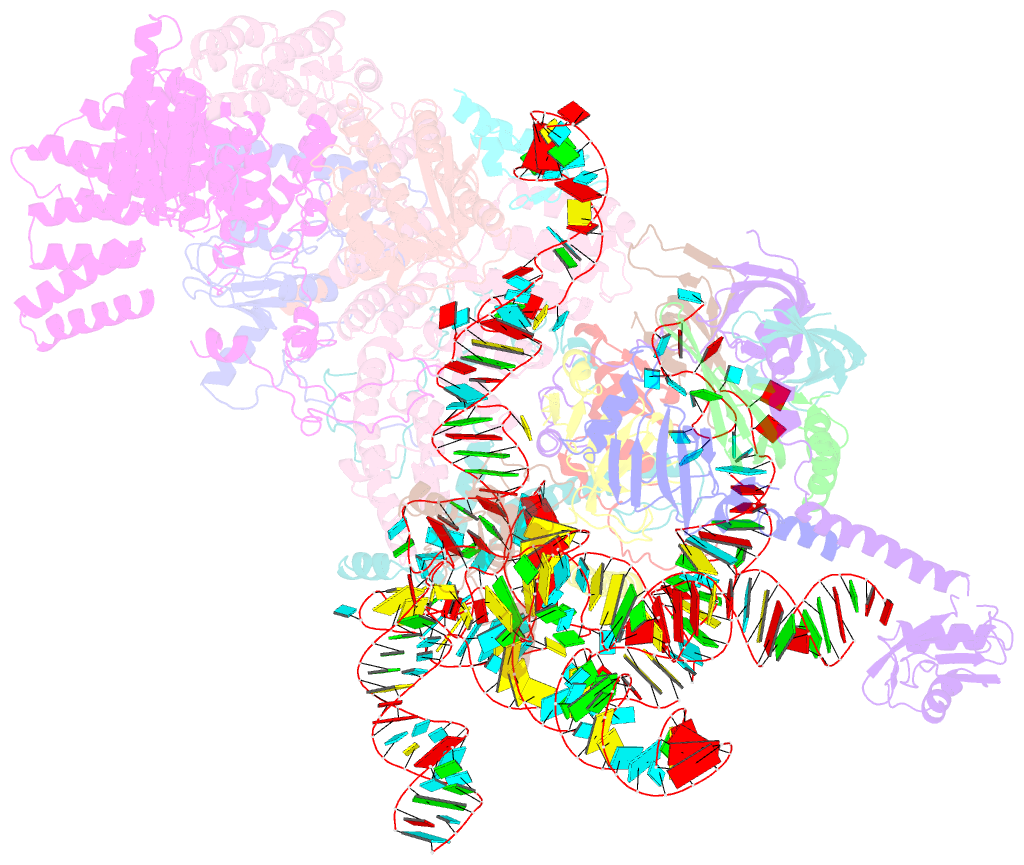
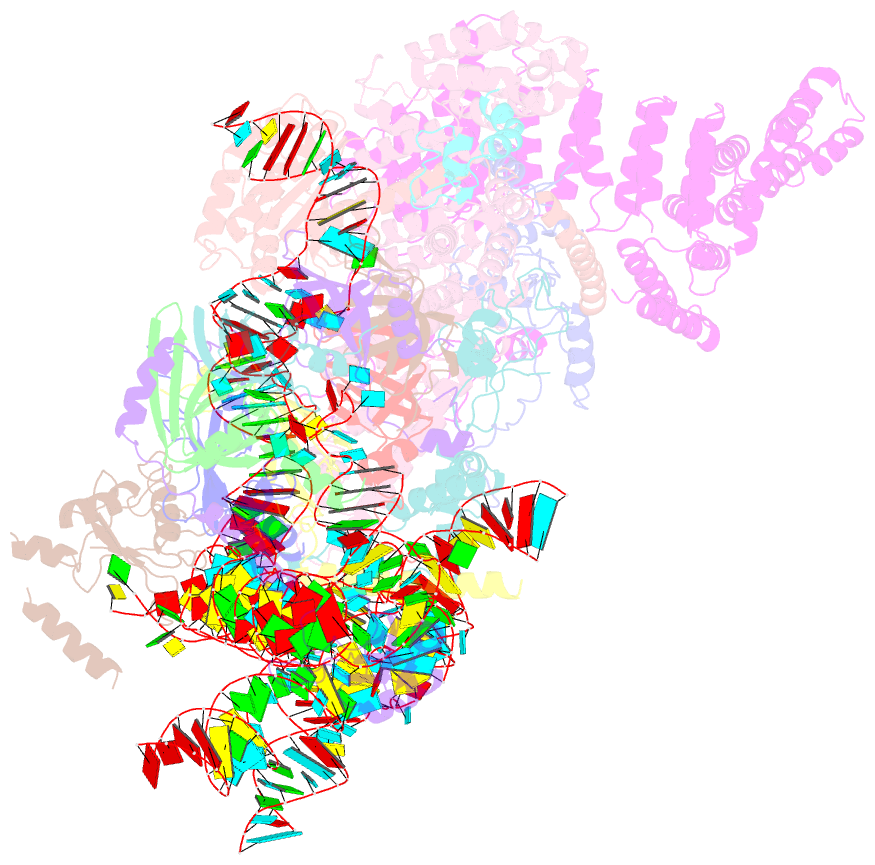
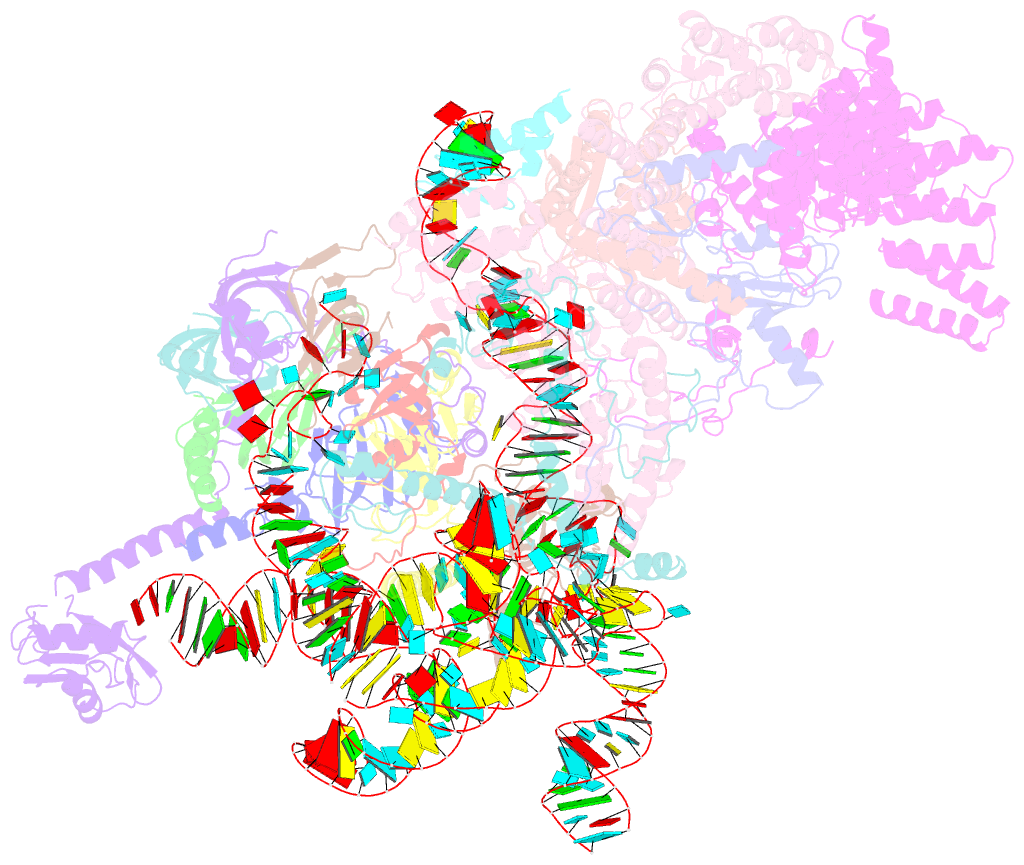
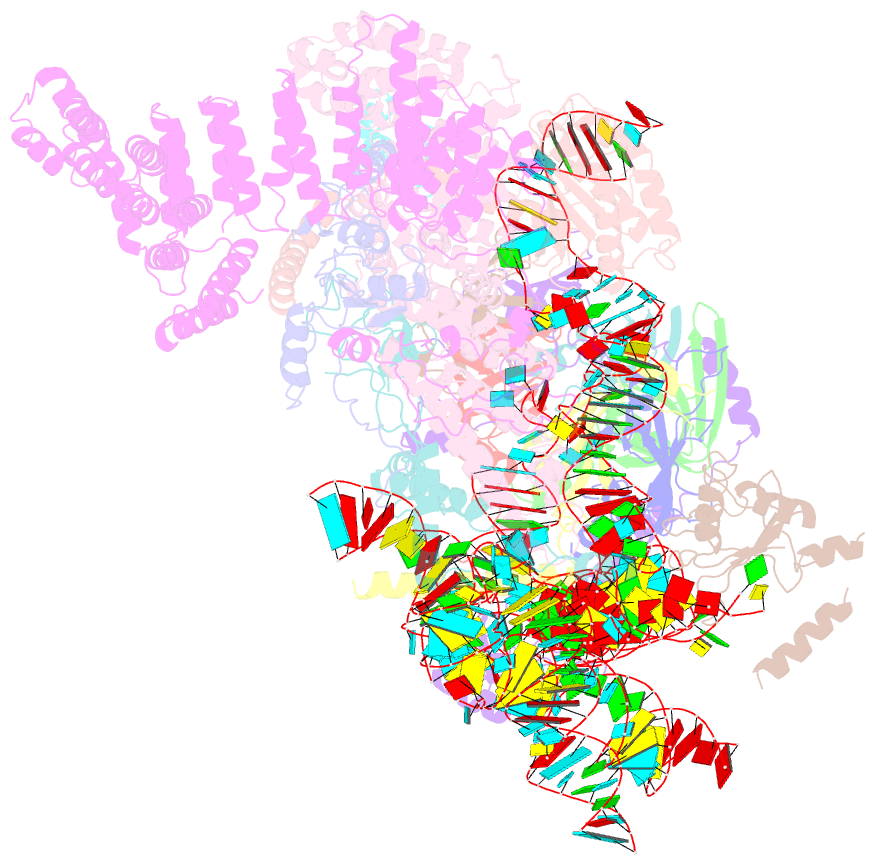
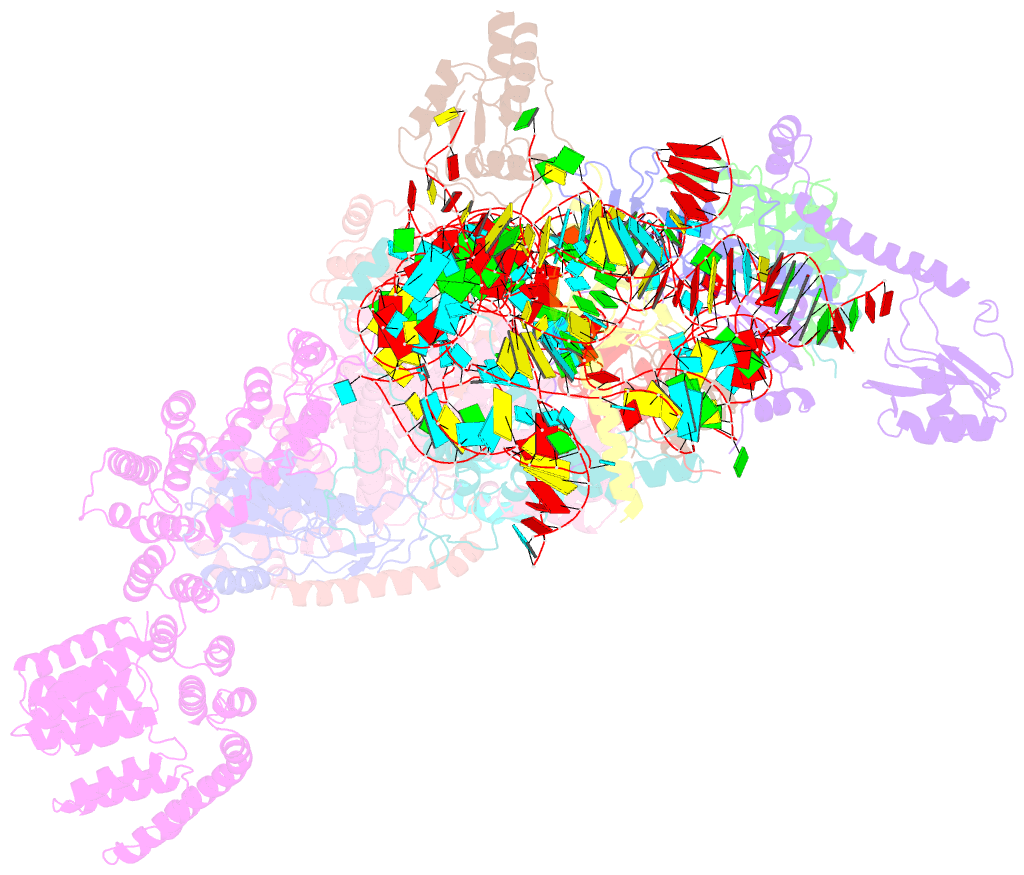
- S. cerevisiae u1 snrnp
- Reference
- Li X, Liu S, Jiang J, Zhang L, Espinosa S, Hill RC, Hansen KC, Zhou ZH, Zhao R (2017): "CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing." Nat Commun, 8, 1035. doi: 10.1038/s41467-017-01241-9.
- Abstract
- U1 snRNP plays a critical role in 5'-splice site recognition and is a frequent target of alternative splicing factors. These factors transiently associate with human U1 snRNP and are not amenable for structural studies, while their Saccharomyces cerevisiae (yeast) homologs are stable components of U1 snRNP. Here, we report the cryoEM structure of yeast U1 snRNP at 3.6 Å resolution with atomic models for ten core proteins, nearly all essential domains of its RNA, and five stably associated auxiliary proteins. The foot-shaped yeast U1 snRNP contains a core in the "ball-and-toes" region architecturally similar to the human U1 snRNP. All auxiliary proteins are in the "arch-and-heel" region and connected to the core through the Prp42/Prp39 paralogs. Our demonstration that homodimeric human PrpF39 directly interacts with U1C-CTD, mirroring yeast Prp42/Prp39, supports yeast U1 snRNP as a model for understanding how transiently associated auxiliary proteins recruit human U1 snRNP in alternative splicing.