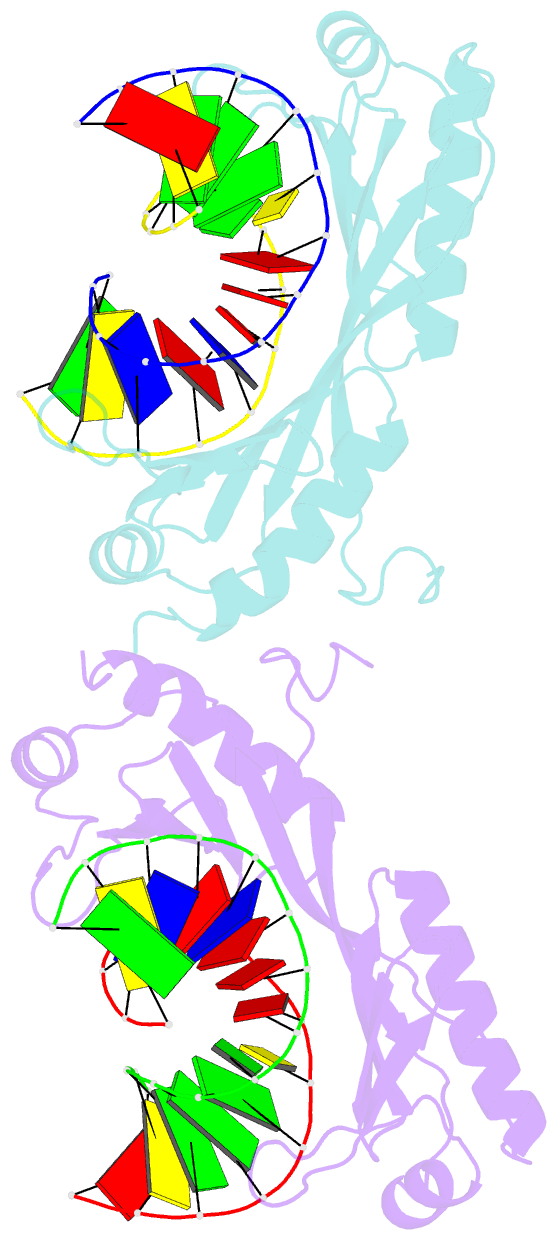
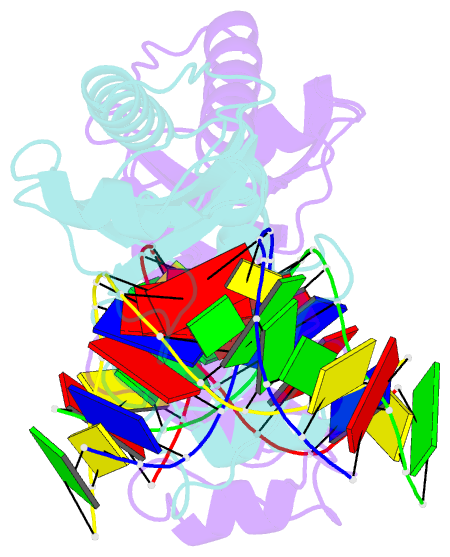
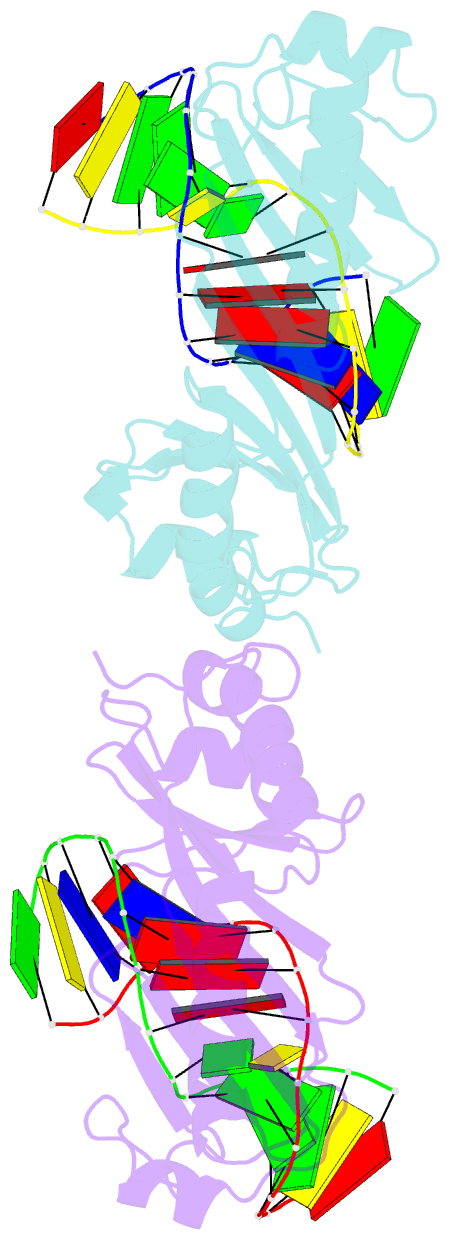
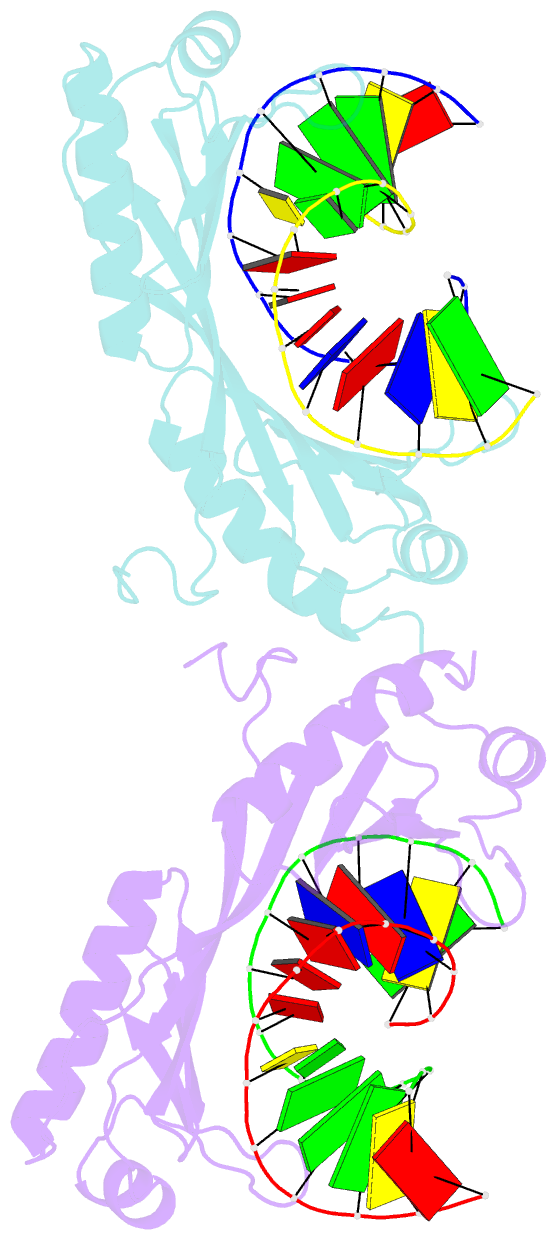
Summary information and primary citation
- PDB-id
- 6njq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.75 Å)
- Summary
- Structure of tbp-hoogsteen containing DNA complex
- Reference
- Stelling AL, Liu AY, Zeng W, Salinas R, Schumacher MA, Al-Hashimi HM (2019): "Infrared Spectroscopic Observation of a G-C+Hoogsteen Base Pair in the DNA:TATA-Box Binding Protein Complex Under Solution Conditions." Angew.Chem.Int.Ed.Engl., 58, 12010-12013. doi: 10.1002/anie.201902693.
- Abstract
- Hoogsteen DNA base pairs (bps) are an alternative base pairing to canonical Watson-Crick bps and are thought to play important biochemical roles. Hoogsteen bps have been reported in a handful of X-ray structures of protein-DNA complexes. However, there are several examples of Hoogsteen bps in crystal structures that form Watson-Crick bps when examined under solution conditions. Furthermore, Hoogsteen bps can sometimes be difficult to resolve in DNA:protein complexes by X-ray crystallography due to ambiguous electron density and by solution-state NMR spectroscopy due to size limitations. Here, using infrared spectroscopy, we report the first direct solution-state observation of a Hoogsteen (G-C+ ) bp in a DNA:protein complex under solution conditions with specific application to DNA-bound TATA-box binding protein. These results support a previous assignment of a G-C+ Hoogsteen bp in the complex, and indicate that Hoogsteen bps do indeed exist under solution conditions in DNA:protein complexes.