Summary information and primary citation
- PDB-id
- 6o16; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.875 Å)
- Summary
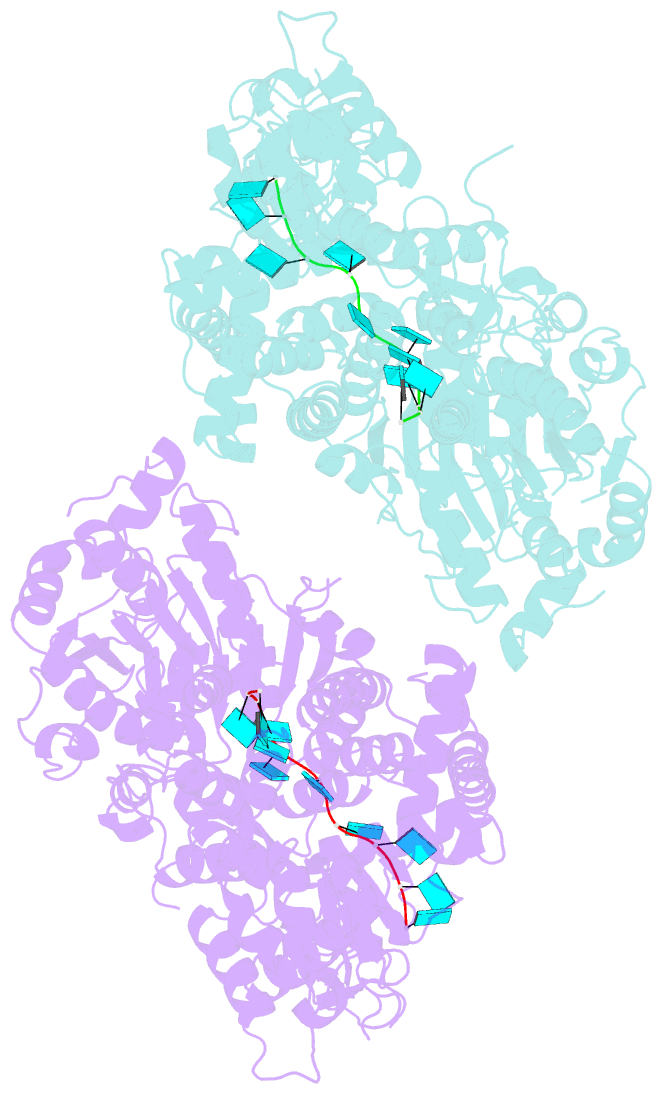
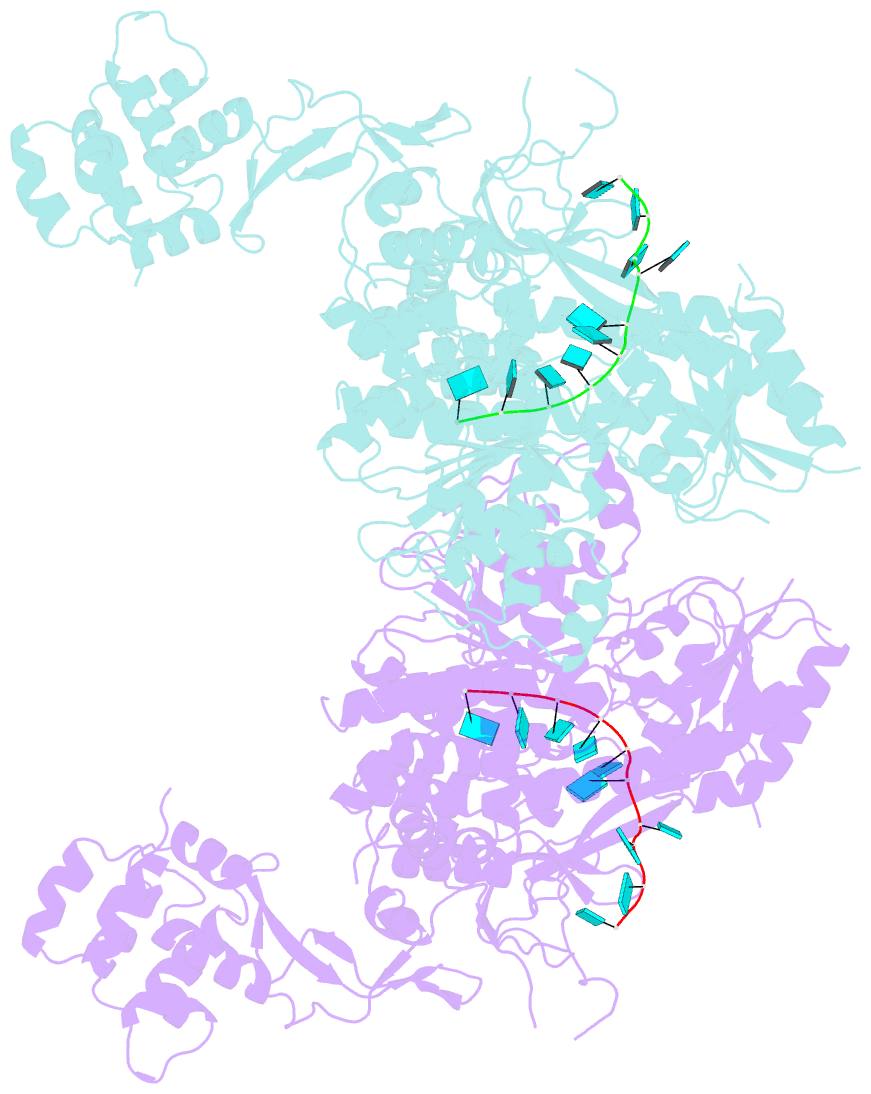
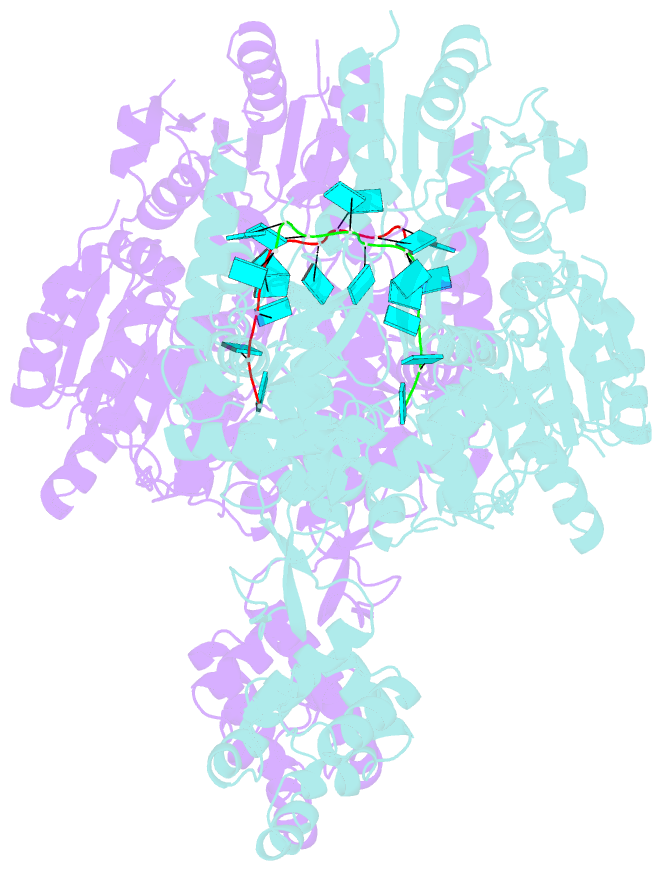
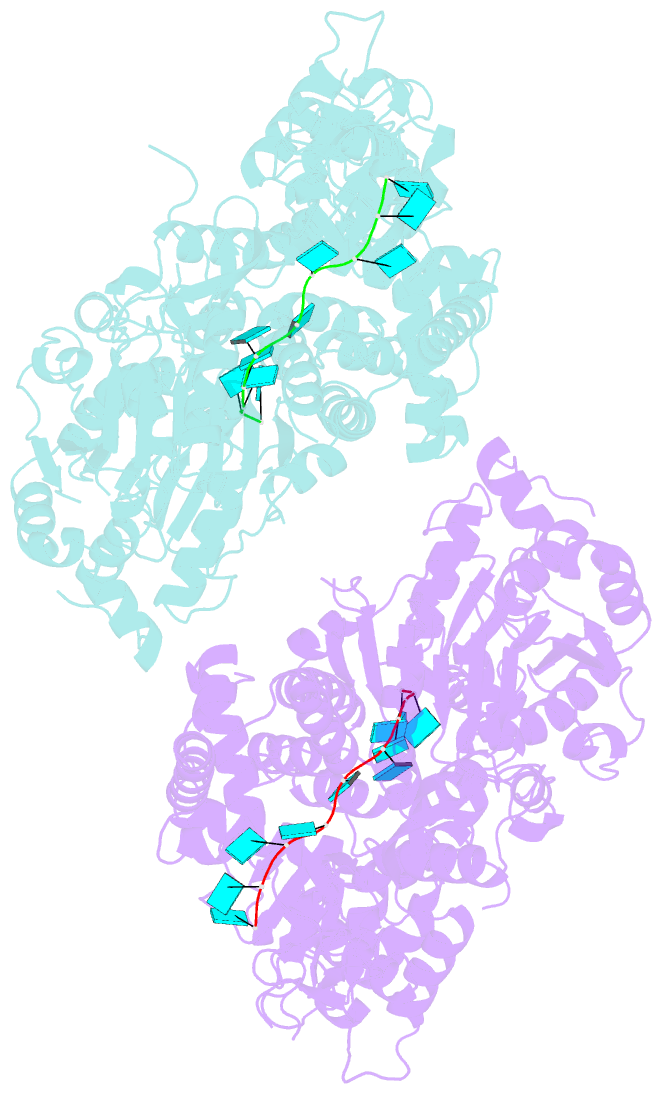
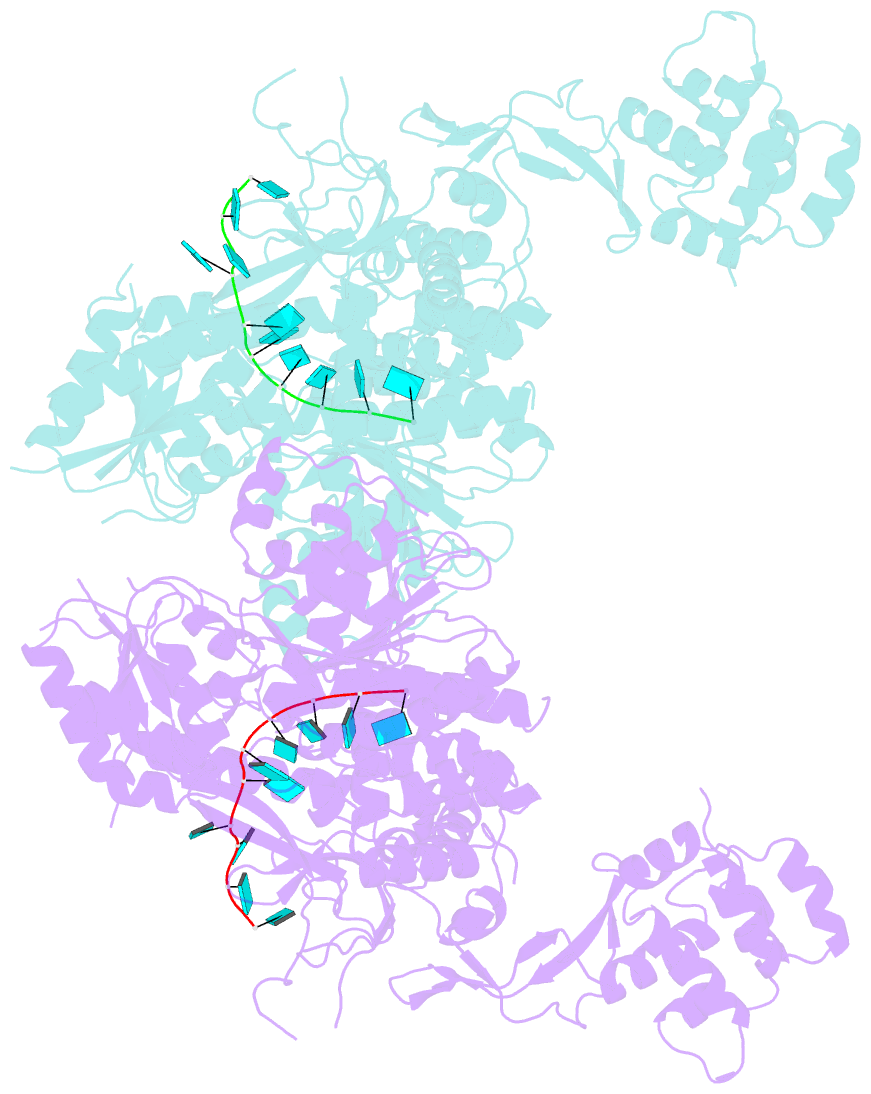
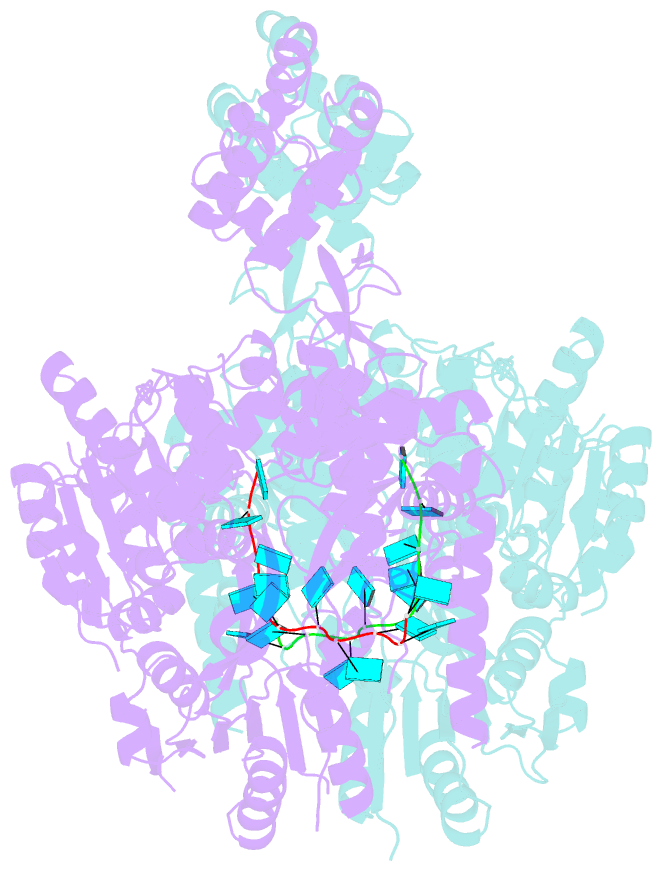
- Crystal structure of murine dhx37 in complex with RNA
- Reference
- Boneberg FM, Brandmann T, Kobel L, van den Heuvel J, Bargsten K, Bammert L, Kutay U, Jinek M (2019): "Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis." Rna, 25, 685-701. doi: 10.1261/rna.069609.118.
- Abstract
- Eukaryotic ribosome biogenesis is a highly orchestrated process involving numerous assembly factors including ATP-dependent RNA helicases. The DEAH helicase DHX37 (Dhr1 in yeast) is activated by the ribosome biogenesis factor UTP14A to facilitate maturation of the small ribosomal subunit. We report the crystal structure of DHX37 in complex with single-stranded RNA, revealing a canonical DEAH ATPase/helicase architecture complemented by a structurally unique carboxy-terminal domain (CTD). Structural comparisons of the nucleotide-free DHX37-RNA complex with DEAH helicases bound to RNA and ATP analogs reveal conformational changes resulting in a register shift in the bound RNA, suggesting a mechanism for ATP-dependent 3'-5' RNA translocation. We further show that a conserved sequence motif in UTP14A interacts with and activates DHX37 by stimulating its ATPase activity and enhancing RNA binding. In turn, the CTD of DHX37 is required, but not sufficient, for interaction with UTP14A in vitro and is essential for ribosome biogenesis in vivo. Together, these results shed light on the mechanism of DHX37 and the function of UTP14A in controlling its recruitment and activity during ribosome biogenesis.