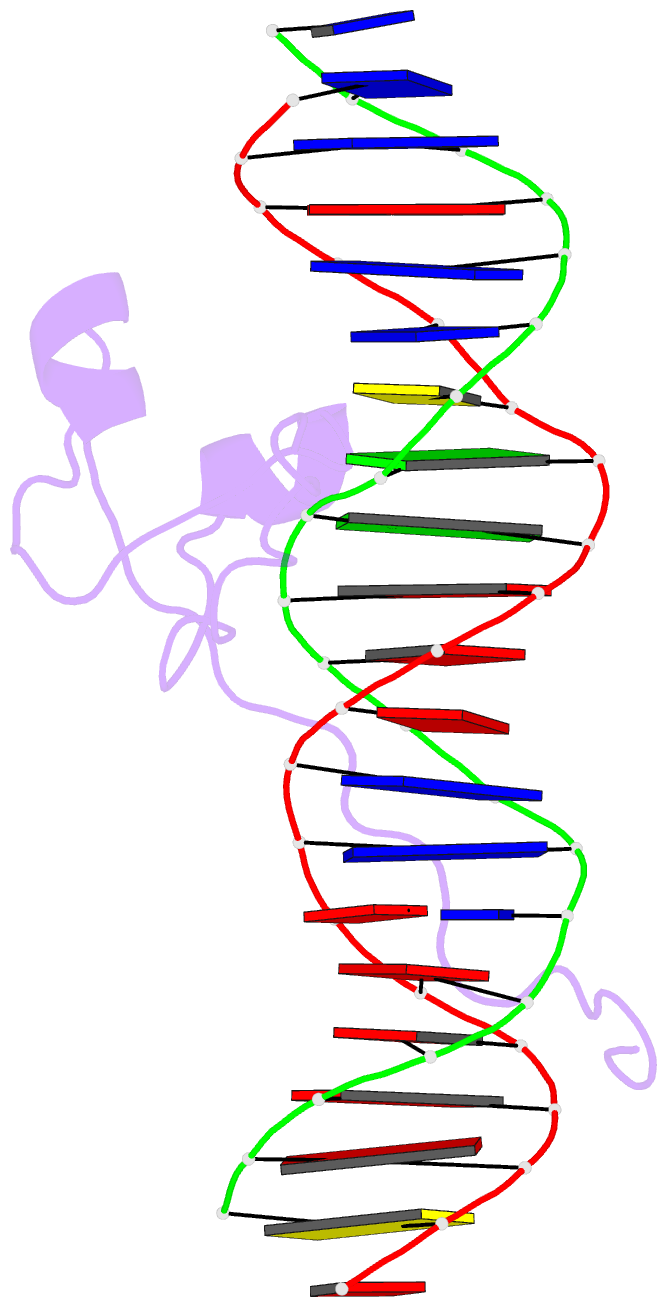
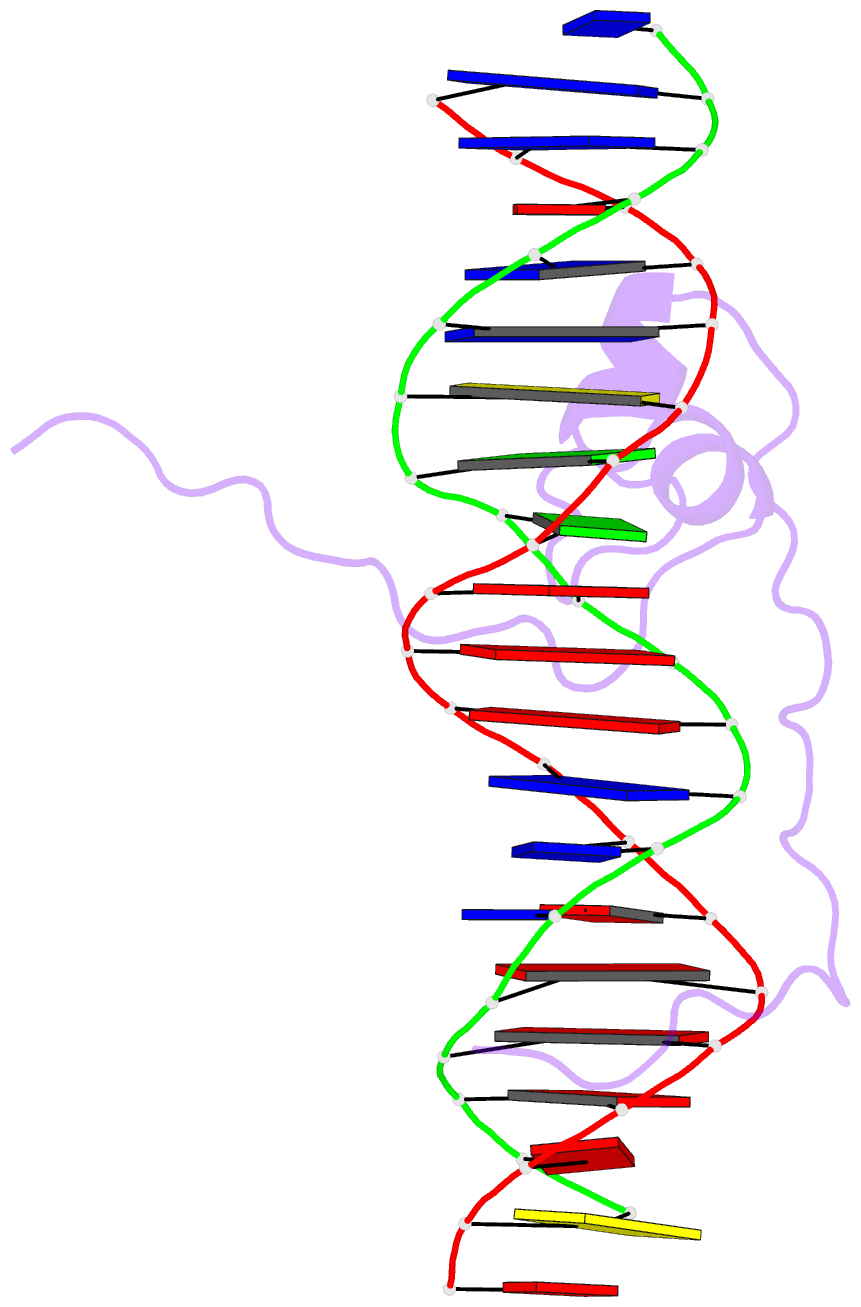
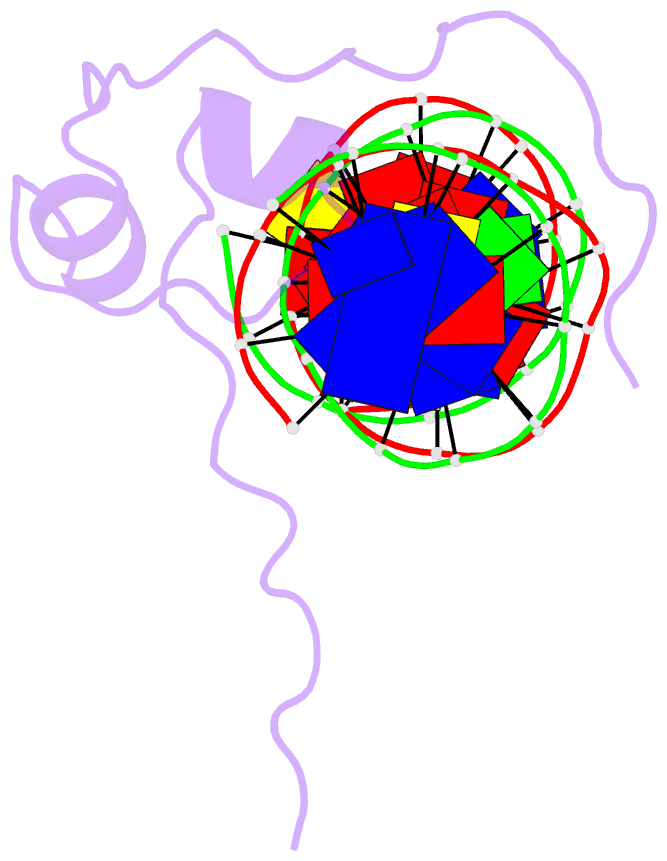
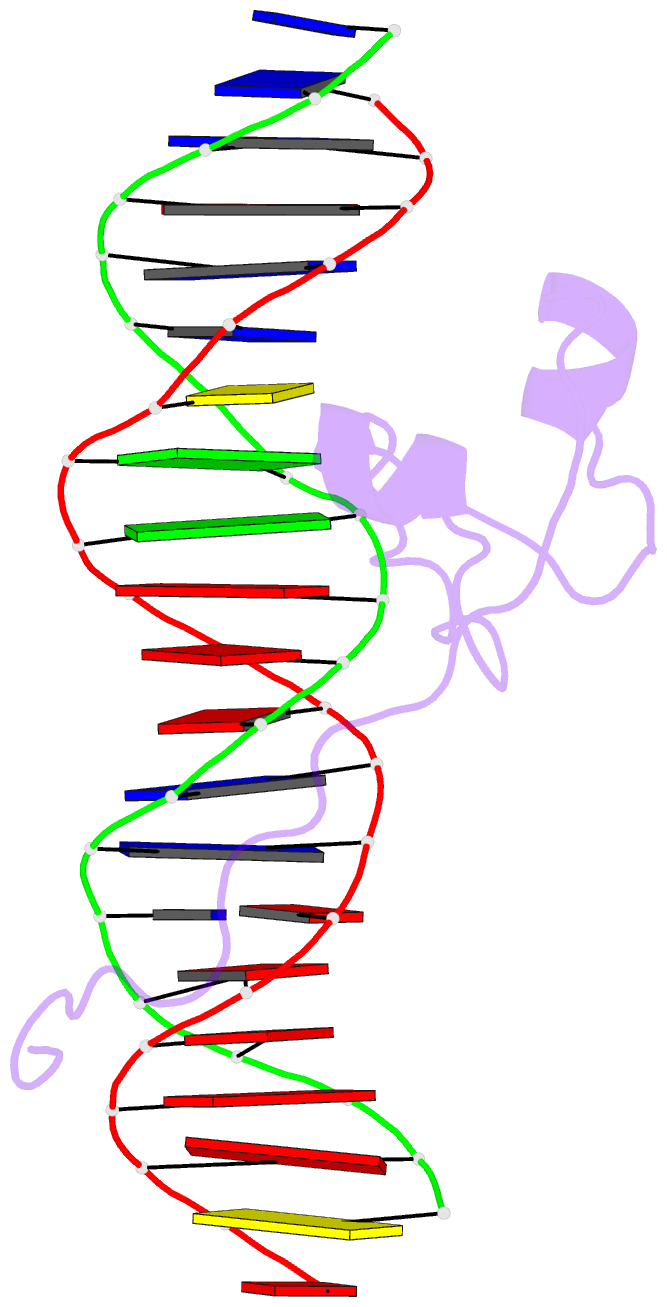
Summary information and primary citation
- PDB-id
- 6o19; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (1.596 Å)
- Summary
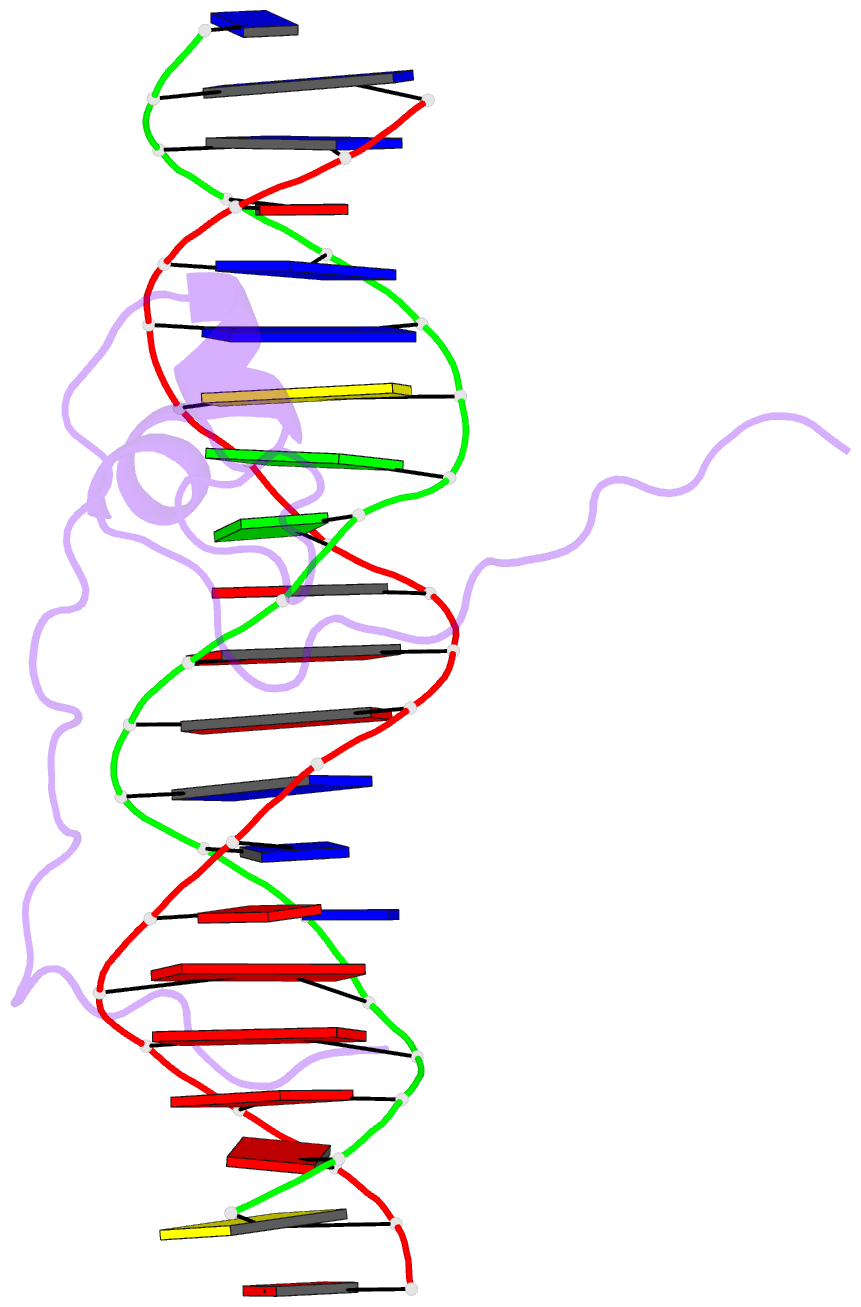
- Crystal structure of pho7 complex with pho1 promoter site 2
- Reference
- Garg A, Goldgur Y, Sanchez AM, Schwer B, Shuman S (2019): "Structure of Fission Yeast Transcription Factor Pho7 Bound topho1Promoter DNA and Effect of Pho7 Mutations on DNA Binding and Phosphate Homeostasis." Mol.Cell.Biol., 39. doi: 10.1128/MCB.00132-19.
- Abstract
- Pho7 is the Schizosaccharomyces pombe fission yeast Zn2Cys6 transcriptional factor that drives a response to phosphate starvation in which phosphate acquisition genes are upregulated. Here we report a crystal structure at 1.6-Å resolution of the Pho7 DNA-binding domain (DBD) bound at its target site 2 in the pho1 promoter (5'-TCGGAAATTAAAAA). Comparison to the previously reported structure of Pho7 DBD in complex with its binding site in the tgp1 promoter (5'-TCGGACATTCAAAT) reveals shared determinants of target site specificity as well as variations in the protein-DNA interface that accommodate different promoter DNA sequences. Mutagenesis of Pho7 amino acids at the DNA interface identified nucleobase contacts at the periphery of the footprint that are essential for the induction of pho1 expression in response to phosphate starvation and for Pho7 binding to site 1 in the pho1 promoter.