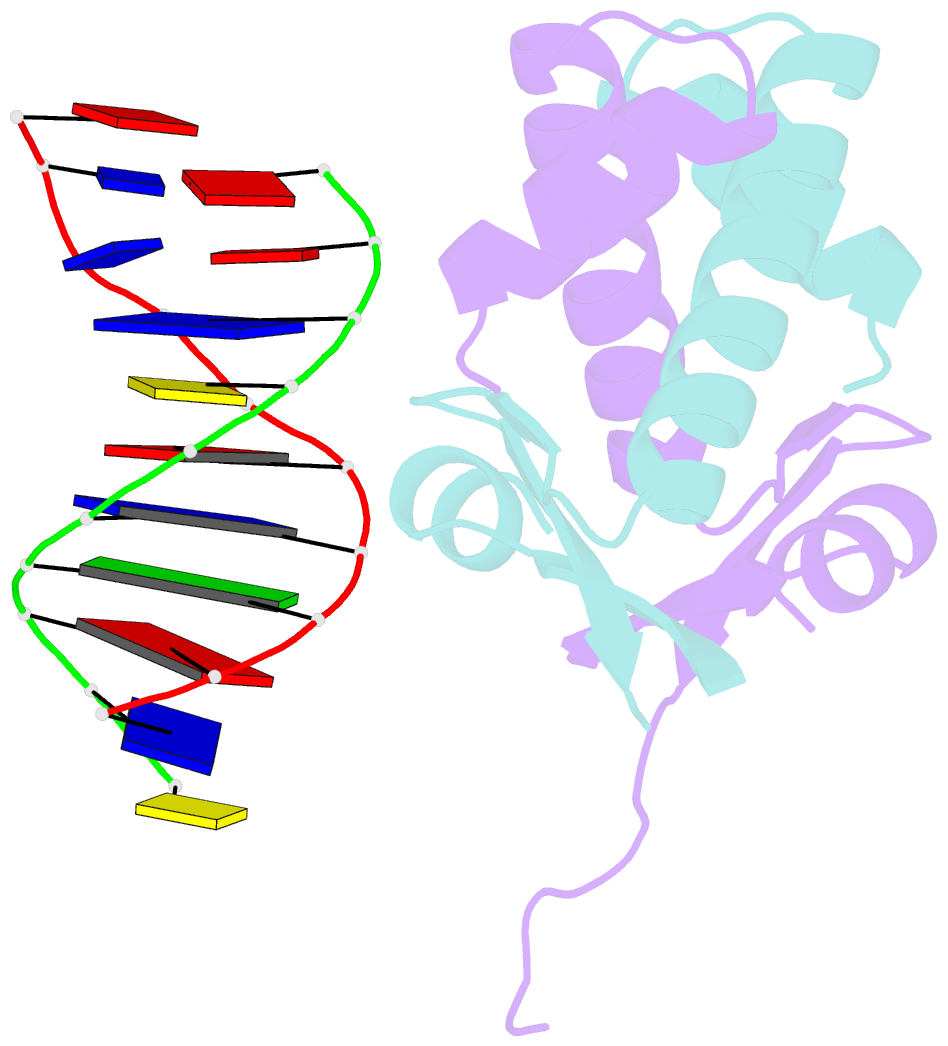
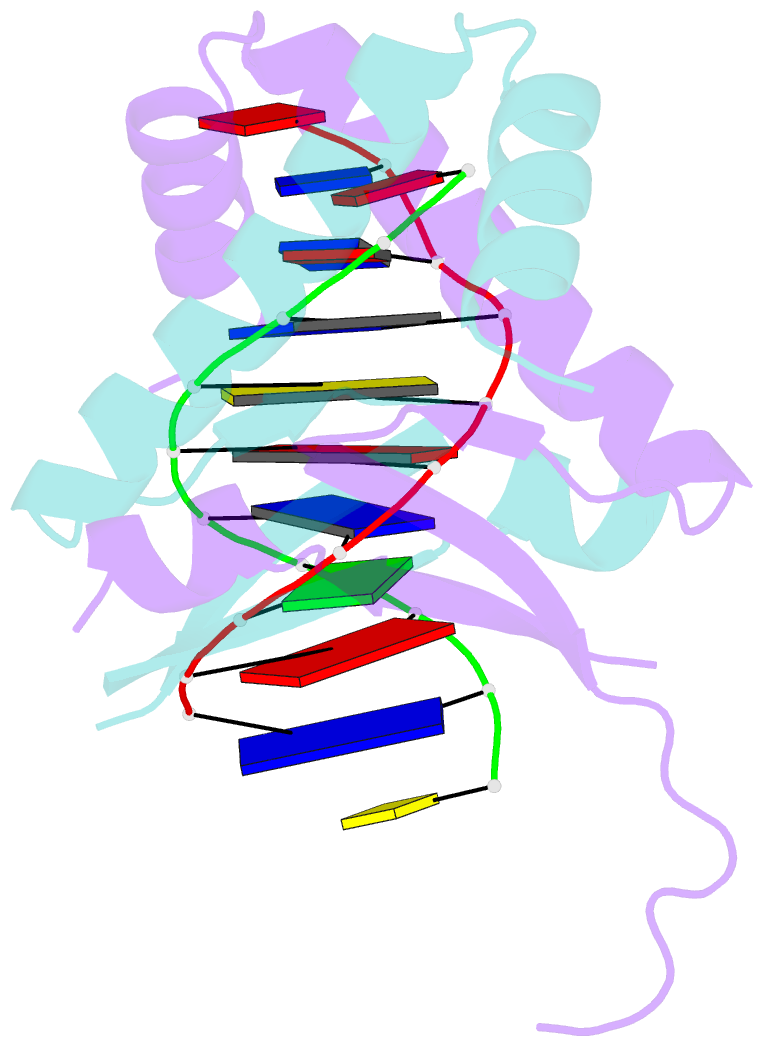
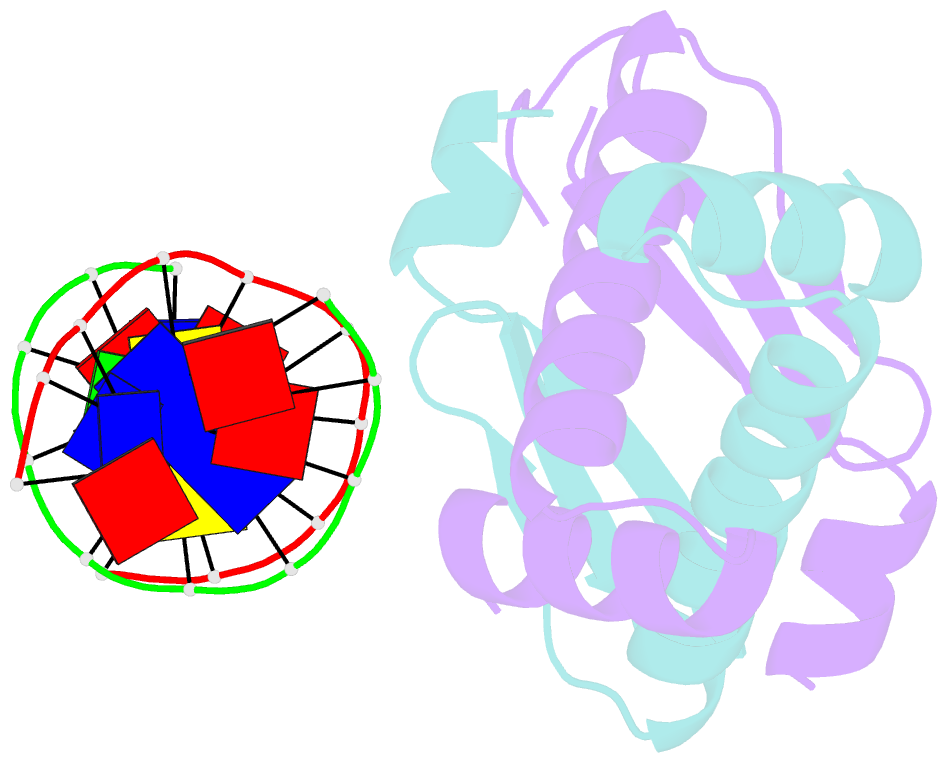
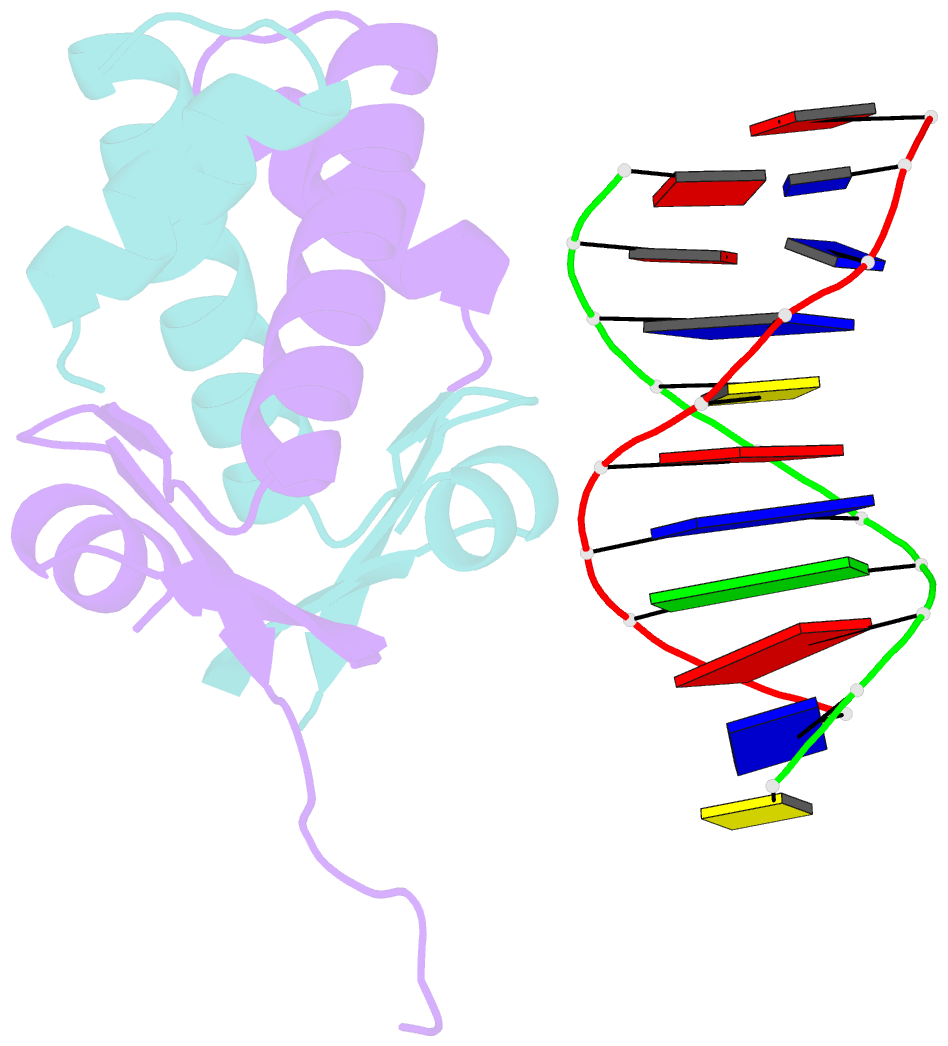
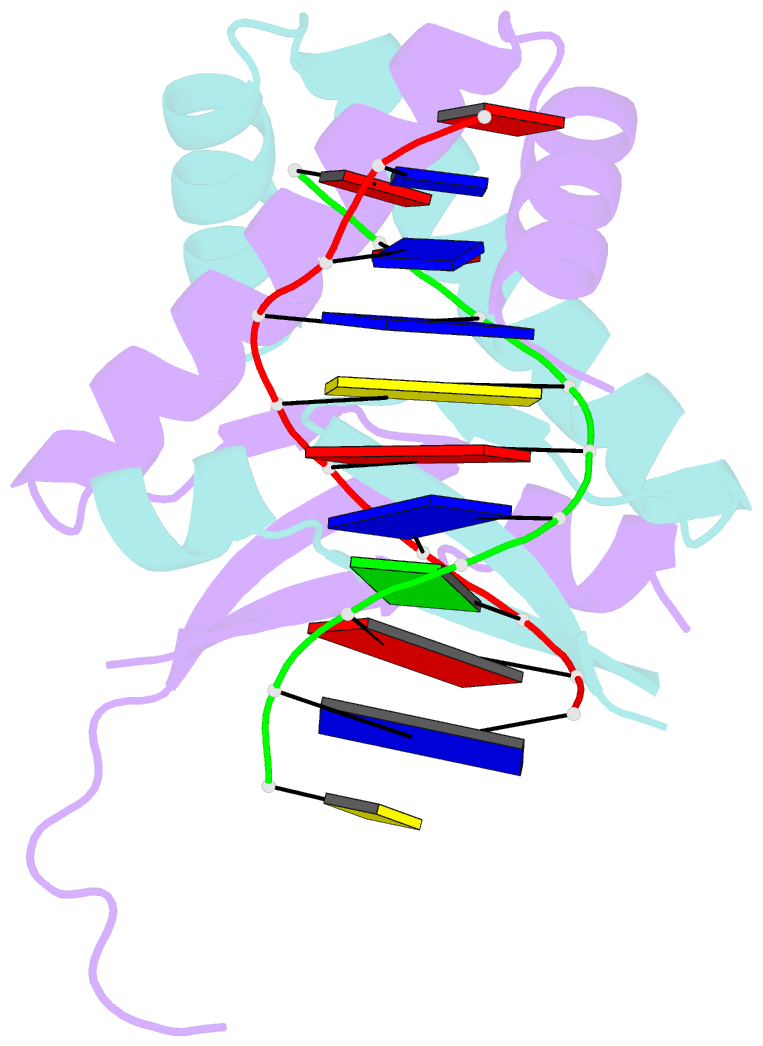
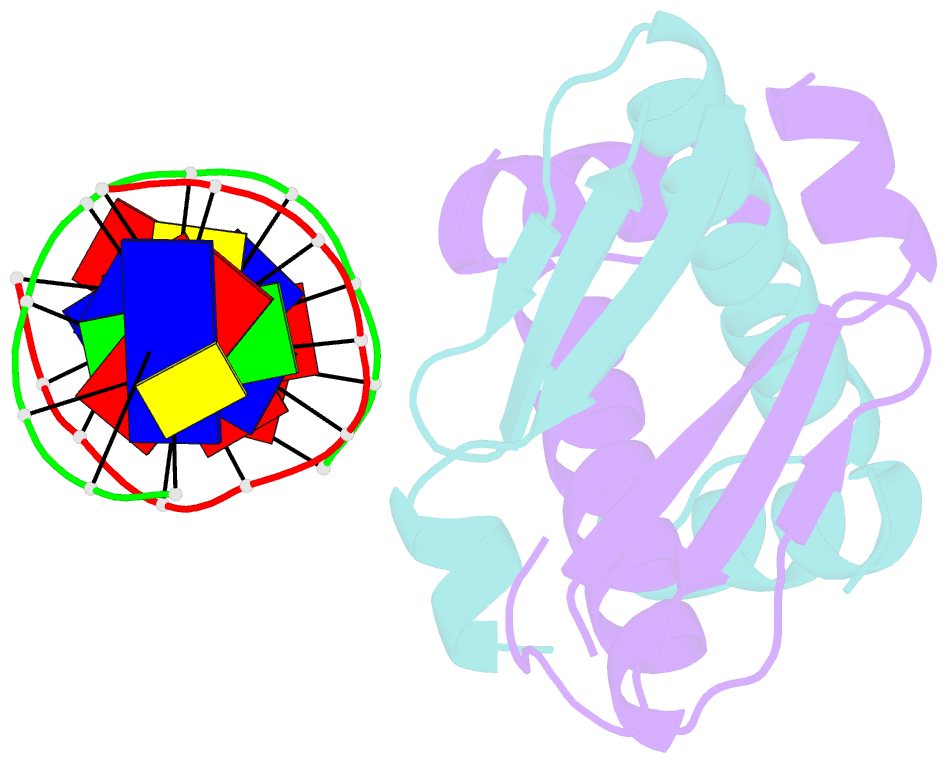
Summary information and primary citation
- PDB-id
- 6o6k; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.601 Å)
- Summary
- Huaa 19bp sym DNA ph 5.5
- Reference
- Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M (2020): "Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling." Nat Commun, 11, 2905. doi: 10.1038/s41467-020-16724-5.
- Abstract
- Bacterial nucleoid remodeling dependent on conserved histone-like protein, HU is one of the determining factors in global gene regulation. By imaging of near-native, unlabeled E. coli cells by soft X-ray tomography, we show that HU remodels nucleoids by promoting the formation of a dense condensed core surrounded by less condensed isolated domains. Nucleoid remodeling during cell growth and environmental adaptation correlate with pH and ionic strength controlled molecular switch that regulated HUαα dependent intermolecular DNA bundling. Through crystallographic and solution-based studies we show that these effects mechanistically rely on HUαα promiscuity in forming multiple electrostatically driven multimerization interfaces. Changes in DNA bundling consequently affects gene expression globally, likely by constrained DNA supercoiling. Taken together our findings unveil a critical function of HU-DNA interaction in nucleoid remodeling that may serve as a general microbial mechanism for transcriptional regulation to synchronize genetic responses during the cell cycle and adapt to changing environments.