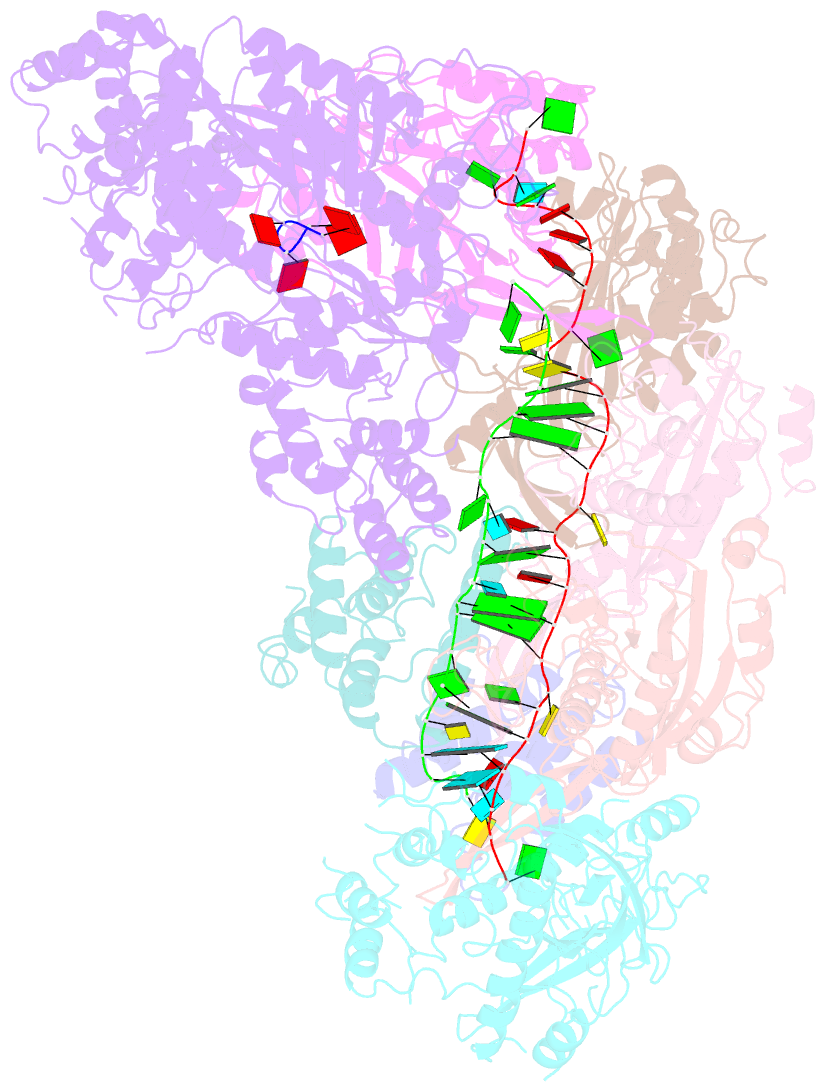
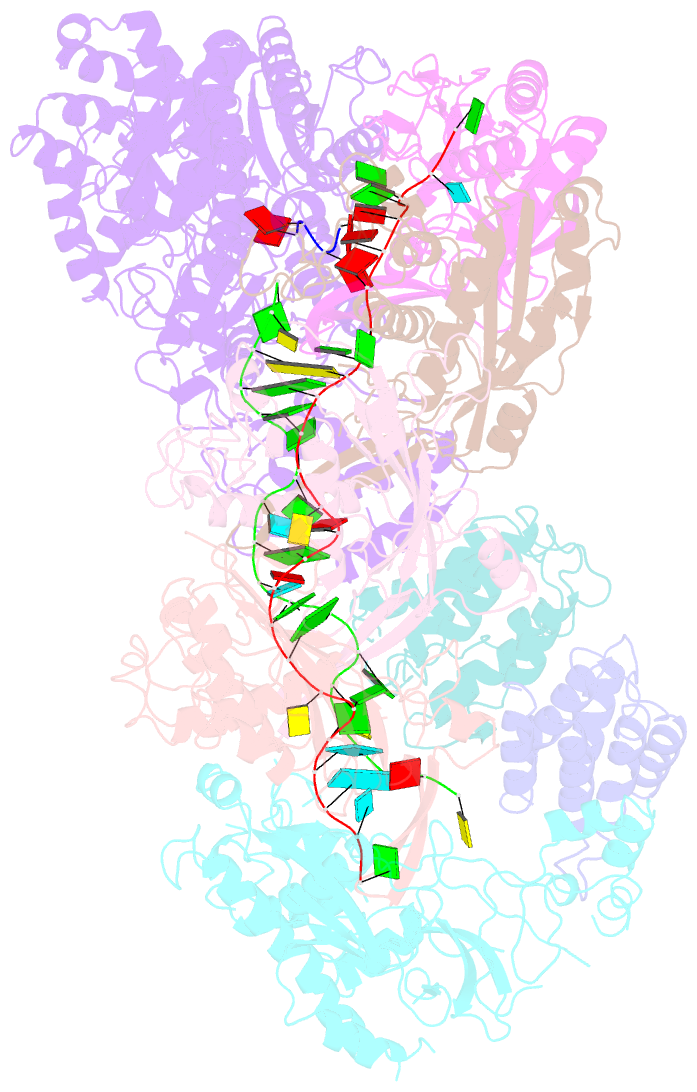
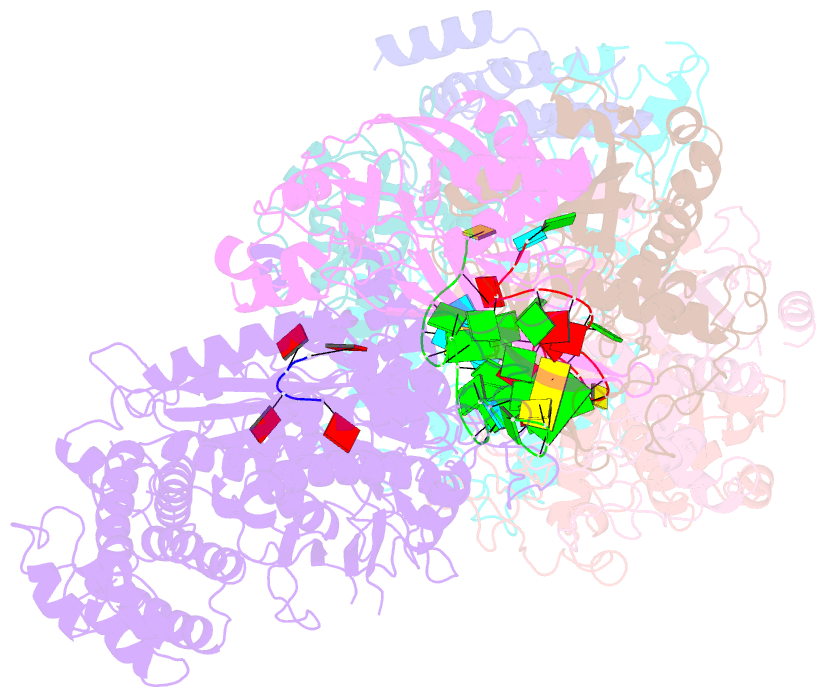
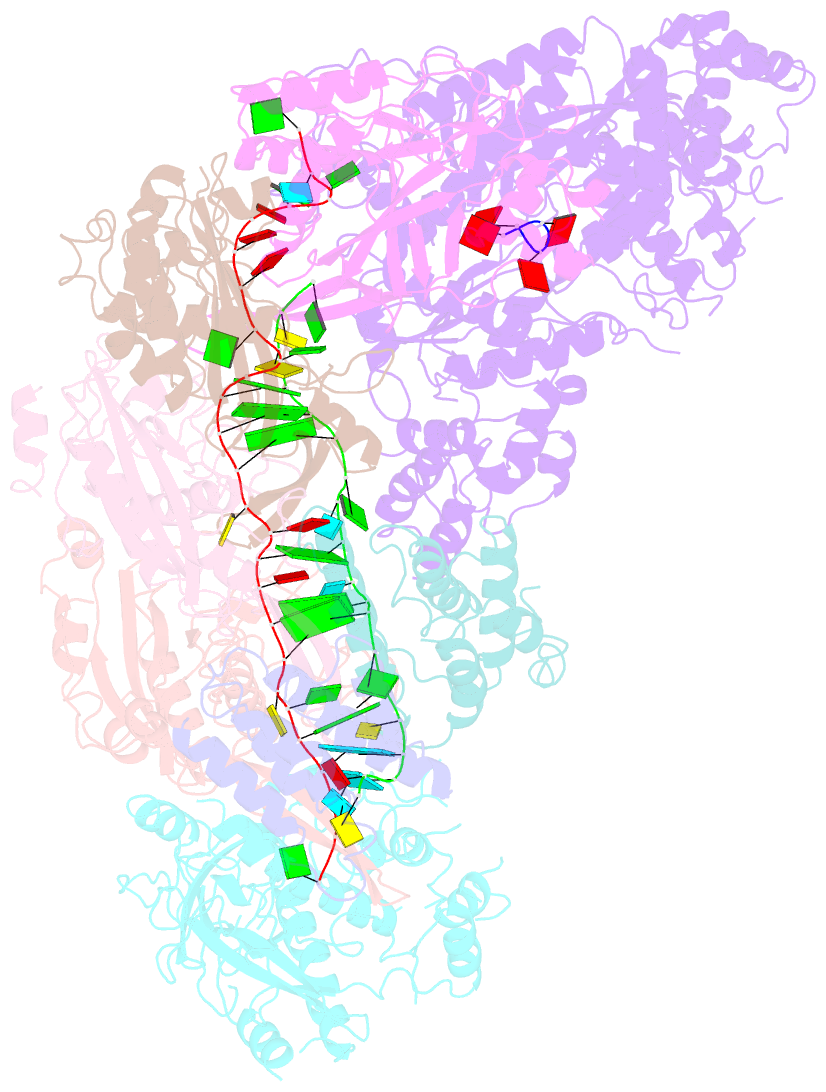
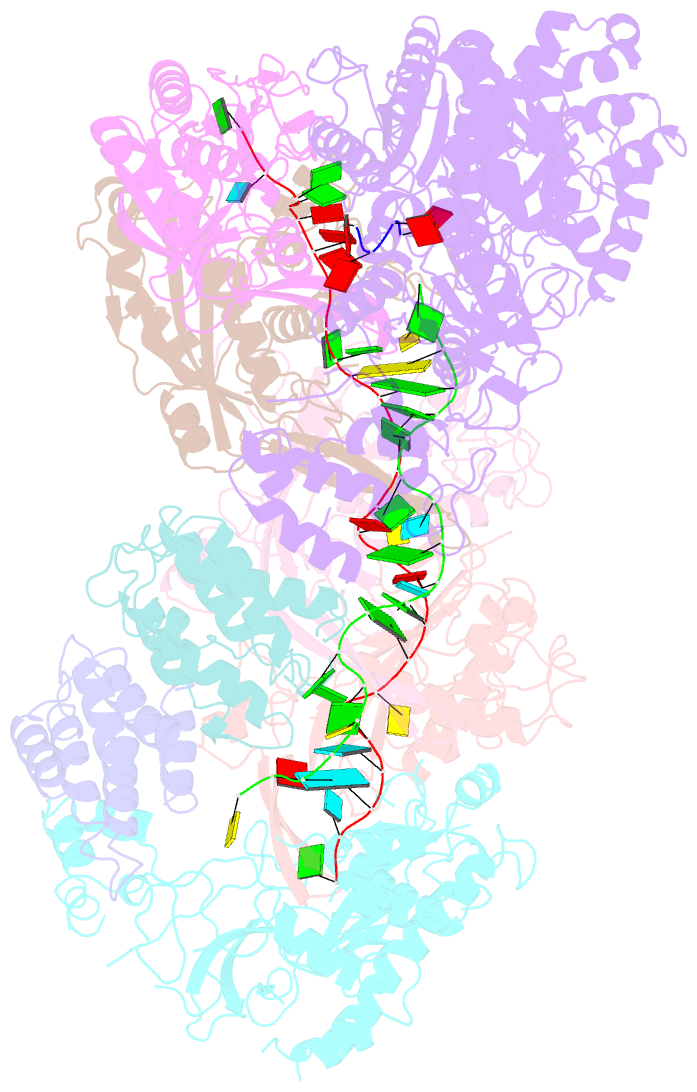
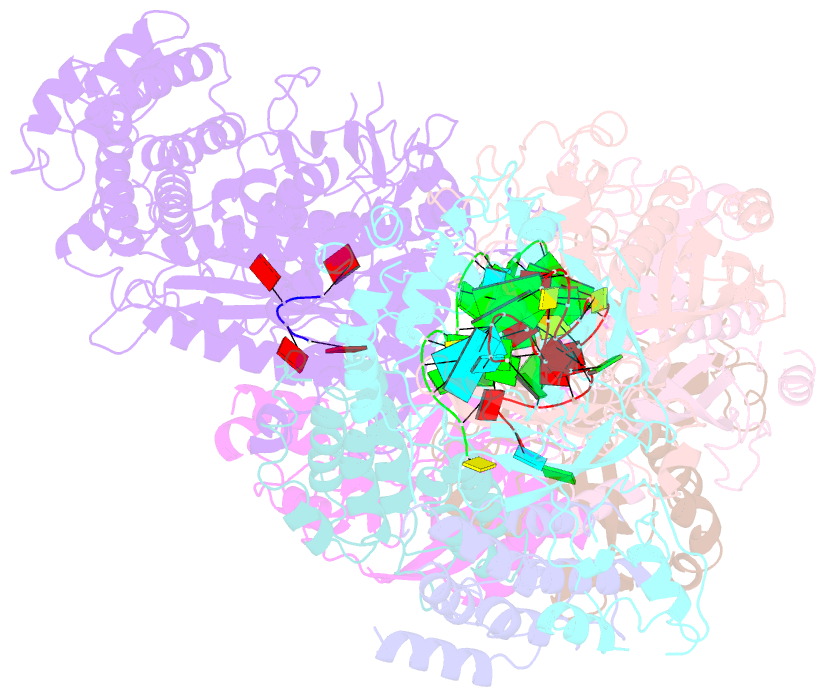
Summary information and primary citation
- PDB-id
- 6o7i; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system-RNA
- Method
- cryo-EM (3.2 Å)
- Summary
- cryo-EM structure of csm-crrna-target RNA ternary bigger complex in complex with ca4 in type iii-a crispr-cas system
- Reference
- Jia N, Jones R, Sukenick G, Patel DJ (2019): "Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path." Mol.Cell, 75, 933-943.e6. doi: 10.1016/j.molcel.2019.06.013.
- Abstract
- Target RNA binding to crRNA-bound type III-A CRISPR-Cas multi-subunit Csm surveillance complexes activates cyclic-oligoadenylate (cAn) formation from ATP subunits positioned within the composite pair of Palm domain pockets of the Csm1 subunit. The generated cAn second messenger in turn targets the CARF domain of trans-acting RNase Csm6, triggering its HEPN domain-based RNase activity. We have undertaken cryo-EM studies on multi-subunit Thermococcus onnurineus Csm effector ternary complexes, as well as X-ray studies on Csm1-Csm4 cassette, both bound to substrate (AMPPNP), intermediates (pppAn), and products (cAn), to decipher mechanistic aspects of cAn formation and release. A network of intermolecular hydrogen bond alignments accounts for the observed adenosine specificity, with ligand positioning dictating formation of linear pppAn intermediates and subsequent cAn formation by cyclization. We combine our structural results with published functional studies to highlight mechanistic insights into the role of the Csm effector complex in mediating the cAn signaling pathway.