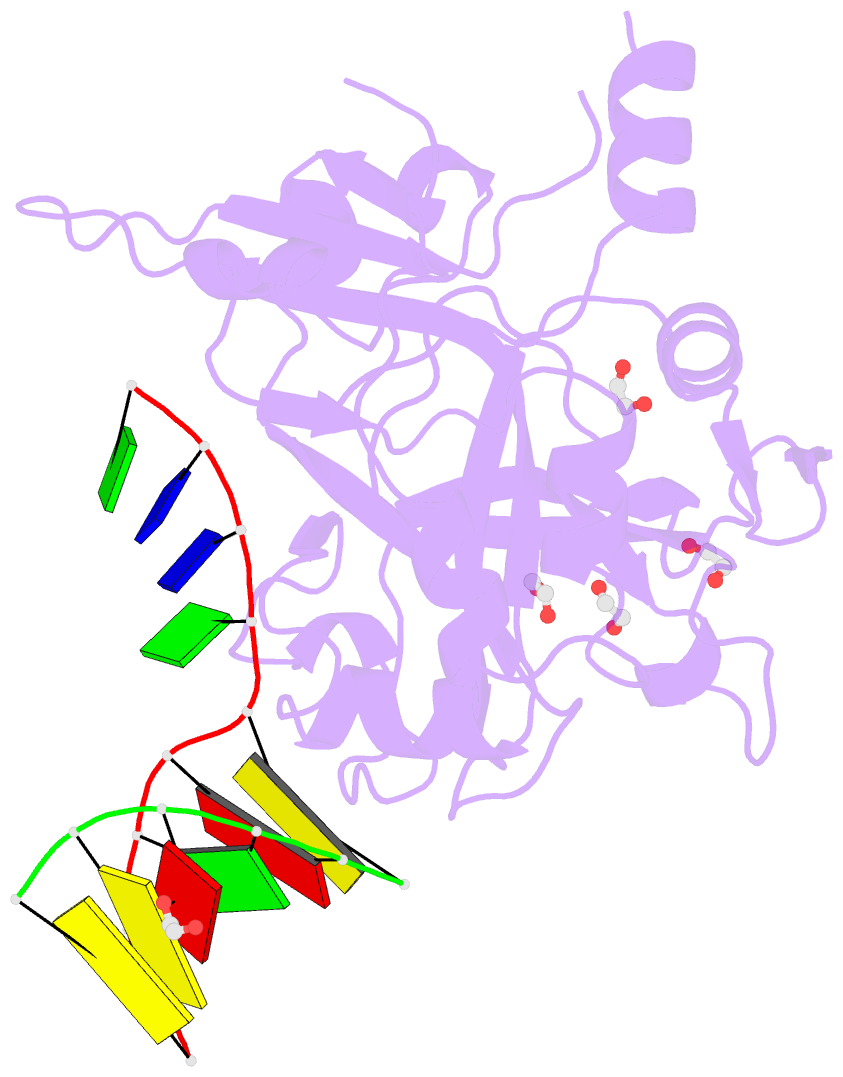
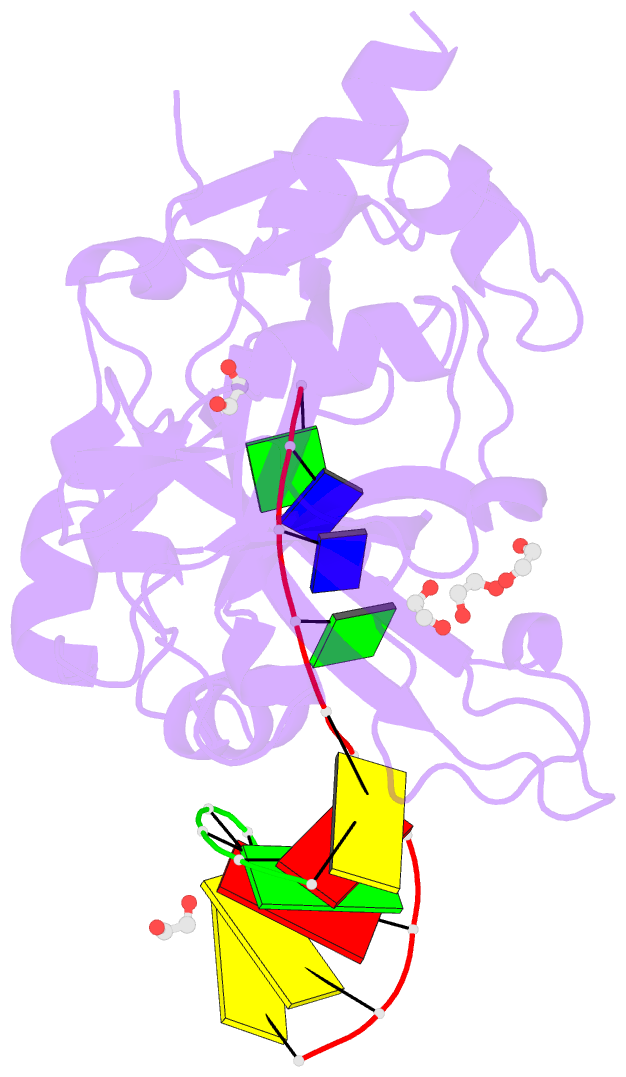
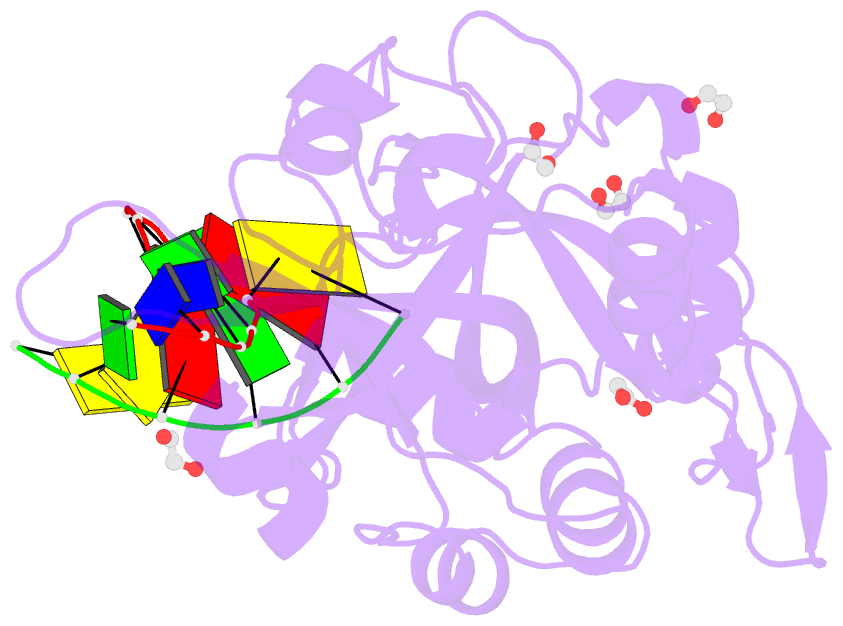
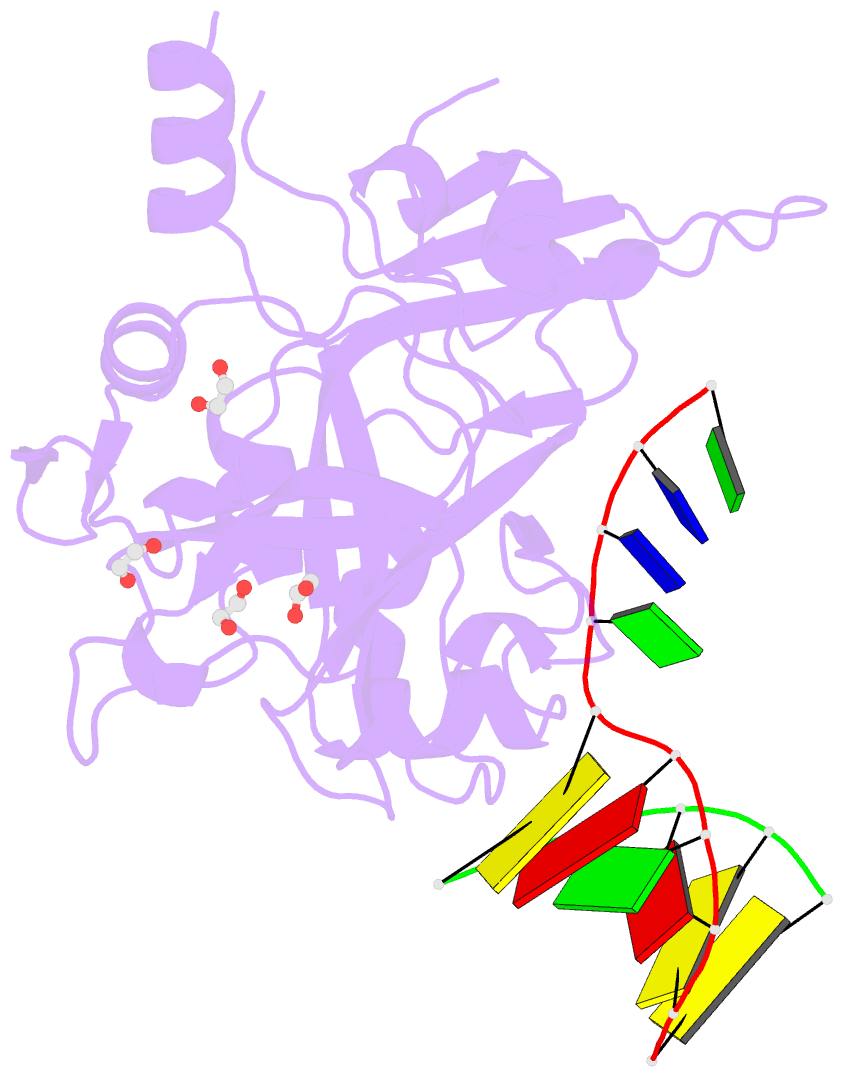
Summary information and primary citation
- PDB-id
- 6oea; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.1 Å)
- Summary
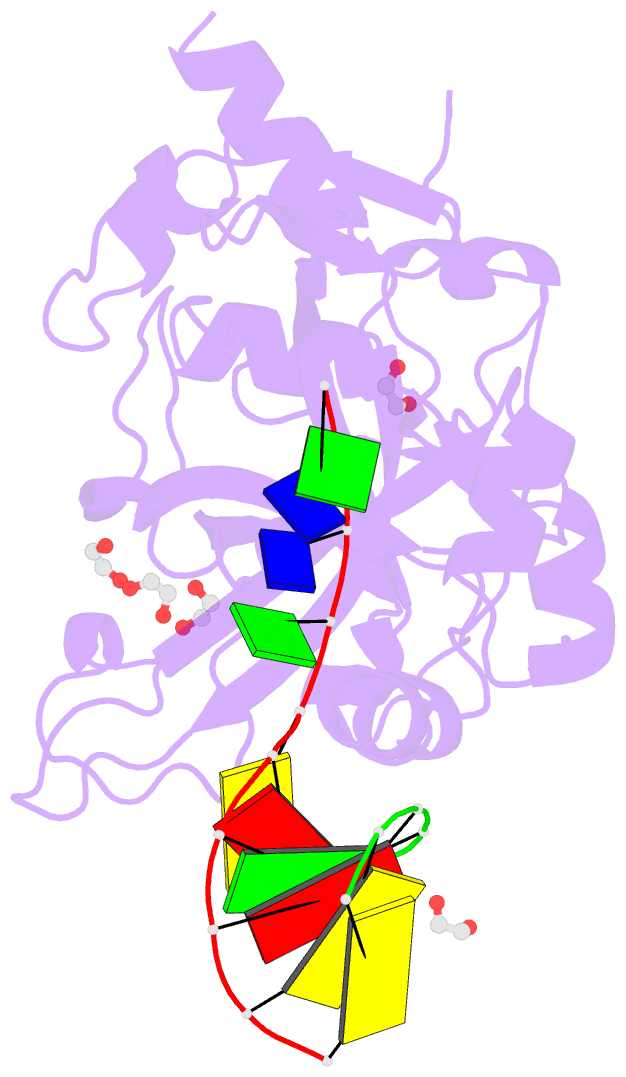
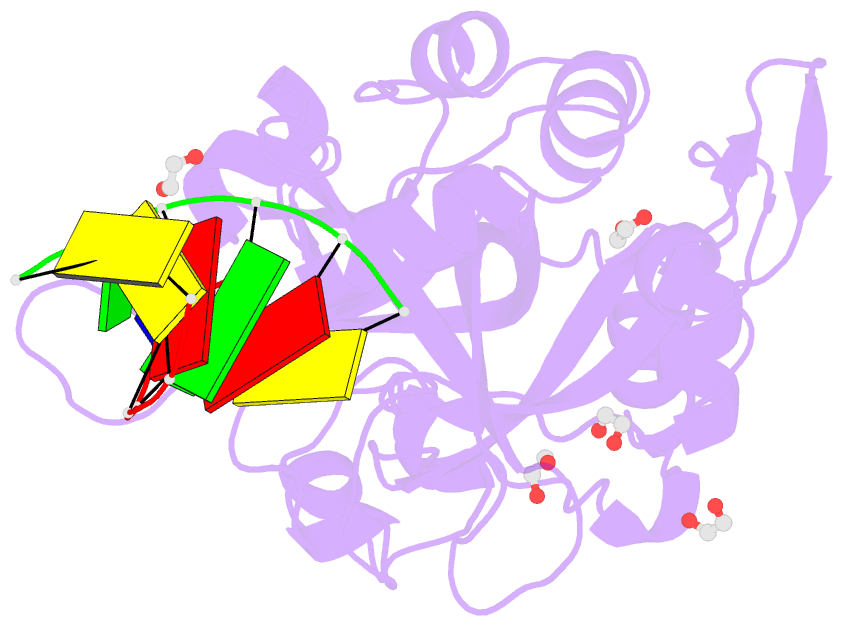
- Crystal structure of hmces srap domain in complex with longer 3' overhang DNA
- Reference
- Halabelian L, Ravichandran M, Li Y, Zeng H, Rao A, Aravind L, Arrowsmith CH (2019): "Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition." Nat.Struct.Mol.Biol., 26, 607-612. doi: 10.1038/s41594-019-0246-6.
- Abstract
- Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein (HMCES) can covalently cross-link to abasic sites in single-stranded DNA at stalled replication forks to prevent genome instability. Here, we report crystal structures of the human HMCES SOS response-associated peptidase (SRAP) domain in complex with DNA-damage substrates, including HMCES cross-linked with an abasic site within a 3' overhang DNA. HMCES interacts with both single-strand and duplex segments of DNA, with two independent duplex DNA interaction sites identified in the SRAP domain. The HMCES DNA-protein cross-link structure provides structural insights into a novel thiazolidine covalent interaction between the DNA abasic site and conserved Cys 2 of HMCES. Collectively, our structures demonstrate the capacity for the SRAP domain to interact with a variety of single-strand- and double-strand-containing DNA structures found in DNA-damage sites, including 5' and 3' overhang DNAs and gapped DNAs with short single-strand segments.